Building of a large-scale, pan-cancer macrophage atlas
Current publications have highlighted the range of TAMs in tumours14,16,17,18. Right here we try to reinforce these efforts by producing a devoted, pan-tumour, single-cell atlas encompassing the total breadth of TAM variety each when it comes to broad-level clustering in addition to an entire catalogue of knowledge on the degree of expression information for particular person genes. We chosen 32 research comprising 17 most cancers varieties16,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48 (Fig. 1a) and processed them utilizing a typical scRNAseq pipeline (Fig. 1b). These information had been obtained via exhaustive searches of each the literature and the Gene Expression Omnibus database. The acquisition technique is detailed in Supplementary Fig. 1. The full dataset contains 363,315 TAMs or macrophage-like cells (i.e. monocytes), with 279,104 cells originating from tumour tissue, 74,982 from adjoining regular, and 9229 cells from different websites (blood, lymph node; Fig. 1c). Of those tumour cells with obtainable annotation, 73.8% originated from main tumour tissue, while 18.5% originated from metastases (the rest had been unknown / NA; Fig. 1d). Lung cancers had the very best variety of cells within the atlas, adopted by clear-cell renal cell carcinomas (ccRCCs) and glioblastoma multiforme (GBM) tumours (Fig. 1e). Extra metadata, resembling histological (e.g. LUAD, LUSC or SCLC for lung most cancers) or molecular subtype (e.g. ER + / HER2+ standing for breast most cancers), for every most cancers was extracted for downstream evaluation.
a Anatomical illustration of the 17 most cancers varieties included within the atlas. b Schematic overview of the methodology of development of the atlas, from information curation, via processing to evaluation. c Pie chart illustrating the distribution of tissue varieties inside the atlas when it comes to cells (proven as numbers). d Pie chart illustrating the proportion of cells originating from main and metastatic tumours included within the atlas when it comes to cells (proven as numbers). e Barplots exhibiting the variety of cells, samples and sufferers per most cancers kind, with color representing the tissue kind (tumour or regular). BCC Basal cell carcinoma; BRCA Breast Most cancers; RCC Renal cell carcinoma; CRC Colorectal most cancers; ESO Esophageal most cancers; GBM Glioblastoma multiforme; LIHC Liver hepatocellular carcinoma; LUAD Lung adenocarcinoma; LUSC lung squamous cell carcinoma; SCLC small cell lung most cancers; MYE Myeloma; OV Ovarian most cancers; PAAD Pancreatic adenocarcinoma; SKCM Pores and skin cutaneous melanoma; THCA Thyroid carcinoma; UCEC Uterine corpus endometrial most cancers; UVM Uveal melanoma. Supply information are offered as a Supply Knowledge file.
The information was generated from a combination of various websites utilizing quite a lot of scRNA sequencing platforms (10x Genomics, MARS-seq, GEXSCOPE, In-Drop and Good-Seq2; Supplementary Fig. 2), in addition to two research using snRNAseq (Supplementary Fig. 3). We carried out batch correction and information integration throughout research earlier than clustering. We benchmarked quite a few approaches utilizing the iLISI criterion49 (strategies) and carried out integration utilizing the RPCA algorithm of Seurat. Clustering and differential expression evaluation had been carried out with native approaches carried out in Seurat. Most cancers/tissue distribution of cells and samples is detailed in Supplementary Knowledge 1, while research/tissue distribution of cells and samples is detailed in Supplementary Knowledge 2. The variety of macrophages and non-macrophages in unique research is detailed in Supplementary Knowledge 3.
The spectrum of TAM variety is broad and complicated
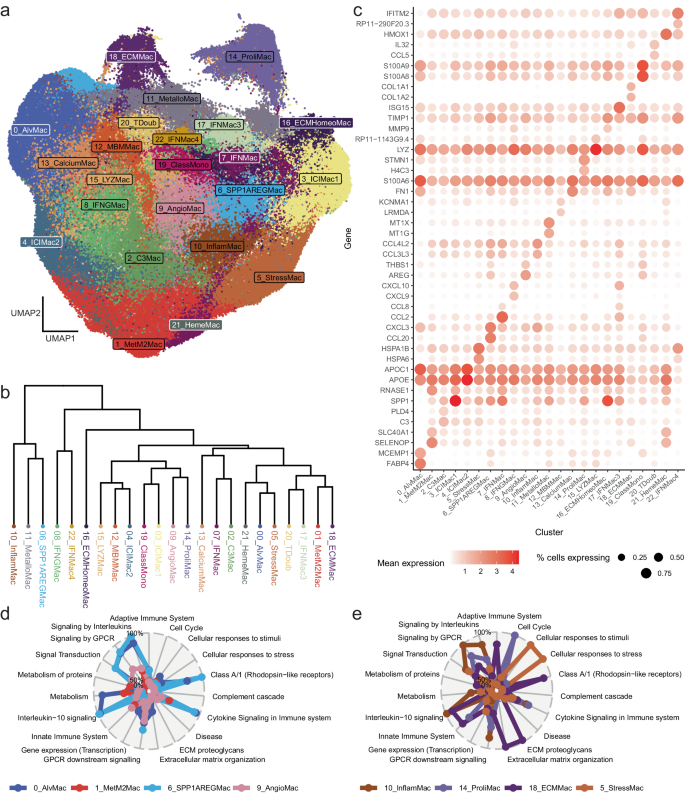
Prevailing fashions for the capabilities of TAMs have superior over time, with current research14 advocating for a transfer away from the normal M1/M2 inflammatory axis classification, to as a substitute give attention to a broader view of those cells that encompasses their numerous phenotypes and capabilities. Right here we recognized TAMs with recurrent phenotypes utilizing a graph-based clustering strategy, which iteratively teams cells collectively utilizing the Louvain algorithm as carried out in Seurat, leading to 23 clusters in whole, visualized as a 2-dimensional UMAP in Fig. 2a. The inter-cluster relationships had been explored utilizing hierarchical clustering (Fig. 2b).
a UMAP visualization of macrophage subsets within the atlas. b Hierarchical clustering evaluation indicating similarity of clusters when it comes to common expression. c Dotplot exhibiting the proportion of cells expressing (measurement) and imply expression (color) of prime 2 most importantly upregulated markers per cluster. d, e Radar plot illustrating key organic pathways upregulated for chosen clusters. Supply information are offered as a Supply Knowledge file.
To evaluate the validity of our clustering strategy, we carried out mapping of identified markers of variety in TAMs14 to every of the clusters, which confirmed right restoration of identified macrophage subsets, resembling IFN-stimulated or proliferating macrophages. As well as, given the well-powered nature of the dataset we recognized uncommon and fewer well-documented subsets, most certainly pertaining to subsets that haven’t beforehand been recognized. Cluster 0, the biggest of the clusters with 38,071 TAMs in whole, represents alveolar macrophages, the vast majority of these TAMs originating from lung tissue, with excessive expression of alveolar macrophage markers FABP4 (Fig. 2c), MCEMP1 and CD5230,50. Cluster 1 was discovered to have immunoregulatory perform, with upregulation of SELENOP, a selenium transporter beforehand related to M2 macrophage polarization51,52; SLC40A1, a part of ferroportin, a mobile iron transporter wherein excessive expression has been proven to advertise M2-polarization of TAMs53, in addition to different M2-associated genes together with PLTP54, F13A155 and FUCA255. Cluster 21 TAMs additionally specific SLC40A1 and SELENOP, however as well as, extremely upregulate CD163 and HMOX1, suggesting that these are heme-clearance macrophages.
Clusters 2, 6, 7, 8, 10 and 17 had been all discovered to be related to irritation, with cluster 2 upregulating C3, required for opsonization and phagocytosis56, PLD4, beforehand related to M1 polarization57 in addition to the MHC class II molecule subunits HLA-DPA1 and HLA-DPB1. Cluster 6 upregulates CCL20, which promotes most cancers cell migration and subsequently development / metastasis58,59; CXCL3, which additionally promotes metastasis in pancreatic most cancers60; the pro-inflammatory cytokine IL1B, in addition to different chemokines CXCL2 and CXCL8 (Fig. 2c, d). Equally, cluster 7 is related to cytokines together with CCL2, CCL8, CCL4L2, CCL3L3, and SPP1, which has been proven to each promote M2 polarization in lung adenocarcinoma61 in addition to being related to angiogenesis62,63. High upregulated genes in cluster 8 embody CXCL9, CXCL10, MMP9, which is required for ECM reworking and subsequently macrophage migration64 and can also be implicated in priming premetastatic websites in lung metastases65, in addition to VAMP5, which is an interferon-induced gene66. Cluster 17 TAMs additionally show an interferon-induced phenotype, the very best upregulated gene being ISG15, in addition to chemokines CXCL10 and CCL8. Equally, cluster 22 TAMs present proof of publicity to interferons, with upregulation of IFITM2 and LST1. Cluster 10 TAMs upregulate quite a lot of cytokines, together with CCL3L3, CCL4L2, CXCL8, IL1B, TNF, CCL4 and CCL3.
There’s important curiosity within the therapeutic focusing on of TAMs as a result of their immunosuppressive perform67, and numerous compounds are in medical growth aiming to both deplete, repolarise or block TAM subset exercise. Cluster 3 TAMs had been present in most most cancers varieties, and partially recapitulated a gene signature that has been related to immunotherapy resistance in melanoma68, with excessive expression of SPP1, RNASE1, NUPR1 and TREM2. TREM2 has been described in numerous contexts, together with within the upkeep of microglial health within the context of Alzheimer illness69, in affiliation with lipid-associated TAMs70, and moreover, experimental information on mice reveals that inhibition of TREM2 potentiates immunotherapy response and inhibits tumour development71. Like cluster 3, TAMs in cluster 4 upregulate TREM2, nonetheless in distinction, in addition they upregulate APOE, a fatty acid metabolism gene studied extensively in neurological issues72 and just lately related to macrophage subsets in breast most cancers42, in addition to APOC1, which has been proven as a possible prognostic biomarker for lung most cancers development73 and when inhibited, promotes transformation of M2 polarized TAMs to an M1 state and enhances anti-PD1 immunotherapy in hepatocellular carcinoma74.
Cluster 5 TAMs are characterised by their upregulation of stress-inducible warmth shock transcripts (Fig. 2c, e), that are related to a broad variety of options of most cancers growth75, together with HSPA6, HSPA1B, HSPA1A, DNAJB1, HSPB1, HSPH1, HSPD1, HSP90AA1 and BAG3, which interacts with warmth shock proteins and can also be induced below demanding stimuli76.
A subset of TAMs have been implicated within the promotion of angiogenesis in tumours14,16 — these correspond to cluster 9 in our atlas, which is the one cluster to considerably differentially specific angiogenesis-associated genes VEGFA, VCAN and THBS1. As well as, TAMs from this cluster extremely upregulate two epidermal development elements, AREG, which is concerned in fibroblast migration by way of macrophage-fibroblast interplay77 and EREG, which promotes early most cancers growth78, in addition to cytokine IL1B.
Cluster 19 seems to be composed of classical monocytes, with these cells upregulating various genes typical to this cell kind, together with S100A8, S100A9, S100A12, VCAN, and LYZ18.
Proliferating TAMs upregulate genes related to cell-cycle and DNA replication; these are represented by cluster 14 TAMs in our atlas. This contains upregulation of H4C3 (histone part), TOP2A (a DNA topoisomerase), some cyclin-dependent kinase associated genes together with CDK1, CDKN3 and CKS1B, in addition to CENPF (kinetochore part), STMN1 (concerned in cell cytoskeleton), and was the one cluster to upregulate MKI67, a canonical marker of proliferation.
For a few of the TAM clusters recognized, their perform with relation to tumour growth and development was much less clear, revealing potential avenues for future experimental elucidation. Cluster 11 TAMs upregulate metallothioneins MT1G, MT1X, MT2A, MT1E, MT1H, MT1F and MT1M, concerned in zinc metabolism and likewise established as a prognostic biomarker for some cancers79,80. Cluster 13 TAMs upregulated FN1, launched within the cargo of extracellular vesicles by TAMs81 and influencive of PDAC response to chemotherapy and M2 polarization in HNSCC82, in addition to S100A6, S100A10, S100A4, all of that are calcium binding proteins. S100A4 acts each intracellularly and extracellularly, the latter has been proven to advertise irritation and metastasis, and is often launched in response to emphasize83. Cluster 15 TAMs had been characterised by upregulation of LYZ, an anti-bacterial enzyme that targets the peptidoglycan part of bacterial cell-walls, and can also be regarded as anti-inflammatory in nature84. Apparently, the highest two upregulated genes in cluster 16 TAMs had been MMP9, a matrix metalloproteinase concerned within the breakdown of extracellular matrix85 and prognostic of breast most cancers86, in addition to TIMP1, which is an inhibitor of MMP9, suggesting a homeostatic position for these TAMs in modulating metalloproteinase exercise.
Cells in cluster 20 seem like mislabelled as macrophages. Cells on this cluster expressed genes related to T cells, resembling NKG7, TRBC2 and CD3D, in addition to a B cell-associated gene, IGKC, probably defined by cell doublets. We retain this cluster to be used as a management in downstream analyses.
TAMs from cluster 12 had been extremely enriched in melanoma tumours, particularly in melanoma mind metastases. The very best upregulated gene from the differential expression evaluation was LRMDA, a melanocyte differentiation issue, suggesting an interactive position between these TAMs and the tumour cells. TAMs in Cluster 18 alternatively upregulate genes concerned in extracellular matrix reworking (Fig. 2c, d), together with COL1A2, COL1A1, COL3A1, SPARC, COL6A2, COL6A1, COL6A3, SPARCL1. Cluster distribution by most cancers kind is proven in Supplementary Fig. 4 and Supplementary Knowledge 4. Cluster distribution by tissue kind is proven in Supplementary Fig. 5.
Each tumour-intrinsic and environmental elements form macrophage phenotype
Having produced an in depth characterization of every macrophage cluster when it comes to their differentially expressed genes and pathways, we then sought to look at whether or not TAM composition inside samples, when it comes to the clusters current, differed between varied situations, together with the tissue web site of the tumour, tumour histology, or tumour molecular subtype. On this means, we aimed to untangle the influences of tumour location within the physique from tumour genotype on macrophage composition, by evaluating TAM-composition in main tumours of e.g. colorectal most cancers to colorectal metastases within the liver and first liver tumours, or by evaluating main melanomas within the pores and skin to melanoma mind metastases and first glioblastomas. To do that, we utilized a statistical technique designed to check for variations in proportion of cell kind between samples inside single cell information87.
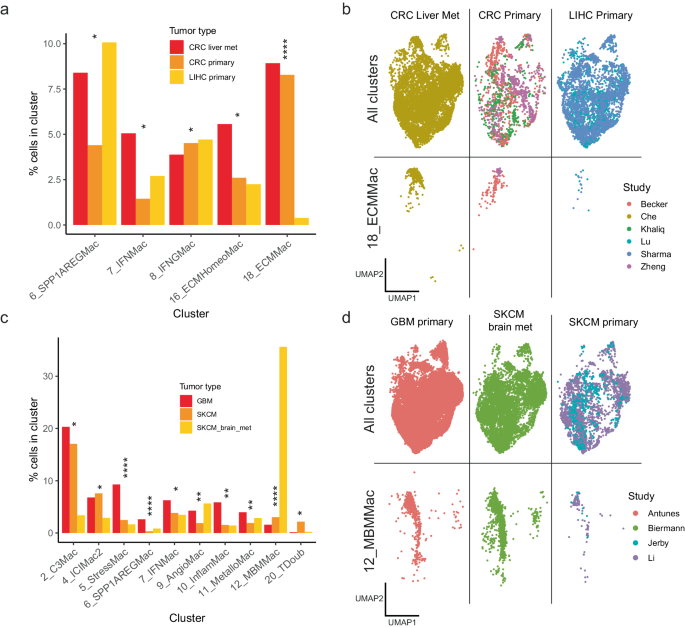
In our comparability of CRC (whether or not main or metastasis; liver metastases derived from ref. 26) and LIHC tumours, we detected important variations within the proportion of 5 clusters between the three situations. Essentially the most outstanding of those variations was within the proportions of cluster 18_ECMMac, which was considerably enriched (moderated two-sided ANOVA by way of propeller87 with FDR correction, q = 0.0000008703) in each CRC primaries and CRC metastases within the liver, however comparatively depleted in main liver tumours (Fig. 3a, b), indicating a possible affect of CRC most cancers cell intrinsic genotypic or phenotypic elements on TAMs subsets. As well as, cluster 6_SPP1AREGMac was considerably enriched (moderated two-sided ANOVA by way of propeller87 with FDR correction, q = 0.02903) in each LIHCs and CRC metastases of the liver however confirmed diminished ranges in main CRCs (Fig. 3a).
a Barplot demonstrating clusters that had been considerably in a different way distributed between main CRCs, metastatic CRCs of the liver, and first LIHCs. The y-axis reveals the proportion of cells from the respective tumour kind belonging to the cluster indicated on the x-axis. Significance testing carried out by way of a moderated two-sided ANOVA utilizing propeller87 with false-discovery fee correction for a number of testing; *q < 0.1, ****q < 0.00005. q-values for 6_SPP1AREGMac, 7_IFNMac, 8_IFNGMac, 16_ECMHomeoMac, 18_ECMMac had been 0.0290377800734, 0.0864189357922, 0.0392092001302, 0.0156655261662 and 0.0000008703445 respectively. b UMAP exhibiting the distribution of TAMs in CRCs and LIHCs, exhibiting all clusters (prime) and 18_ECMMac (backside). CRC Colorectal most cancers, LIHC Liver hepatocellular carcinoma. c Barplot exhibiting clusters considerably in a different way distributed between main melanomas, melanoma metastases within the mind and first glioblastomas. The y-axis reveals the proportion of cells from the respective tumour kind belonging to the cluster indicated on the x-axis. Significance testing carried out by way of a moderated two-sided ANOVA utilizing propeller87 with false-discovery fee correction for a number of testing; *q < 0.1, **q < 0.01 ****q < 0.00005. q-values for 12_MBMMac, 6_SPP1AREGMac, 5_StressMac, 9_AngioMac, 10_InflamMac, 11_MetalloMac, 7_IFNMac, 4_ICIMac2, 20_TDoub, 2_C3Mac, had been 0.000000001027255, 0.0000004568955, 0.0000490144, 0.002526791, 0.005923449, 0.007044681, 0.02982392, 0.02982392, 0.02982392 and 0.02982392 respectively. d UMAP exhibiting the distribution of TAMs within the melanomas and glioblastomas, exhibiting all clusters (prime) and 12_MBMMac solely (backside). GBM Glioblastoma multiforme, SKCM Pores and skin cutaneous melanoma. Supply information are offered as a Supply Knowledge file.
The comparability of melanomas (main or metastasis) and GBMs additionally yielded a number of important variations within the proportions of macrophage subsets (Fig. 3c, d). Most strikingly, cluster 12_MBMMac, wherein the highest differentially expressed gene was LRMDA, a identified melanocyte differentiation issue, was considerably extremely enriched (moderated two-sided ANOVA by way of propeller87 with FDR correction, q = 0.00000000102) in melanoma mind metastases in comparison with each main melanomas and first glioblastomas (Fig. 3c, d), suggestive of an interplay between this tumour genotype and the encompassing mind tissue. As well as, cluster 2_C3Mac was considerably enriched (moderated two-sided ANOVA by way of propeller87 with FDR correction, q = 0.02982) in each main melanomas and first glioblastomas in comparison with melanoma mind metastases.
By way of histology and molecular subtype, we additionally examined for variations in macrophage composition between lung adenocarcinomas (LUAD) and lung squamous cell carcinomas (LUSC), in addition to between breast cancers with various receptor standing, together with HR + , HER2 + , HR + /HER2+ and triple destructive breast cancers (TNBC). This evaluation revealed considerably larger proportions of clusters 16_ECMHomeoMac and 6_SPP1AREGMac in LUSC, and conversely, considerably larger proportions of 15_LYZMac in LUAD (Moderated two-sided T-test by way of Propeller with FDR correction for a number of testing, q = 0.03168, q = 0.00000387, q = 0.08006 respectively; Supplementary Fig. 6). As well as, TNBCs had considerably larger proportions of 14_ProliMac in comparison with HR + /HER2+ optimistic breast cancers, with the reverse pattern for 2_C3Macs, which had been larger in HR + /HER2+ optimistic breast cancers in comparison with TNBCs (Moderated two-sided T-test by way of Propeller with FDR correction for a number of testing, q = 0.0594 and q = 0.0761 respectively; Supplementary Fig. 7). The upper proportion of proliferating macrophages in TNBCs is concordant with the excessive proliferative exercise and elevated immune infiltrate of TNBCs in comparison with different breast cancers88.
TAM phenotypes and affected person responses to immune checkpoint inhibitors
TAMs have lengthy been related to therapeutic end result67,89,90, together with the response to immune checkpoint inhibitors (ICI)91. There’s additionally appreciable curiosity within the repolarization of TAMs from M2 to M1 phenotype92, a method that might affect ICI end result, in addition to utilizing M1 TAMs as drug supply vectors93. We hypothesized that particular macrophage subsets in our atlas is perhaps related to ICI response, and assessed this with an expanded model of our beforehand revealed CPI1000 cohort, right here encompassing 1446 ICI-treated sufferers with bulk RNA-seq information and known as the CPI1000+ cohort94 (strategies).
Firstly, we outlined scRNAseq cluster gene expression signatures primarily based on the highest differentially expressed markers per cluster, and evaluated their potential efficacy in bulk information utilizing a second atlas containing all cell varieties (see strategies, Supplementary Knowledge 5–6). This allowed us to evaluate whether or not the signatures had been macrophage-specific or not. We went on to outline a set of macrophage-specific “gold-standard” signatures, which constantly recognized their respective macrophage clusters when assessed by way of UCell scores within the all cell-type atlas (strategies), particularly for clusters 5_StressMac, 6_SPP1AREGMac, 8_IFNGMac, 11_MetalloMac, 17_IFNMac3, 21_HemeMac and 22_IFNMac4.
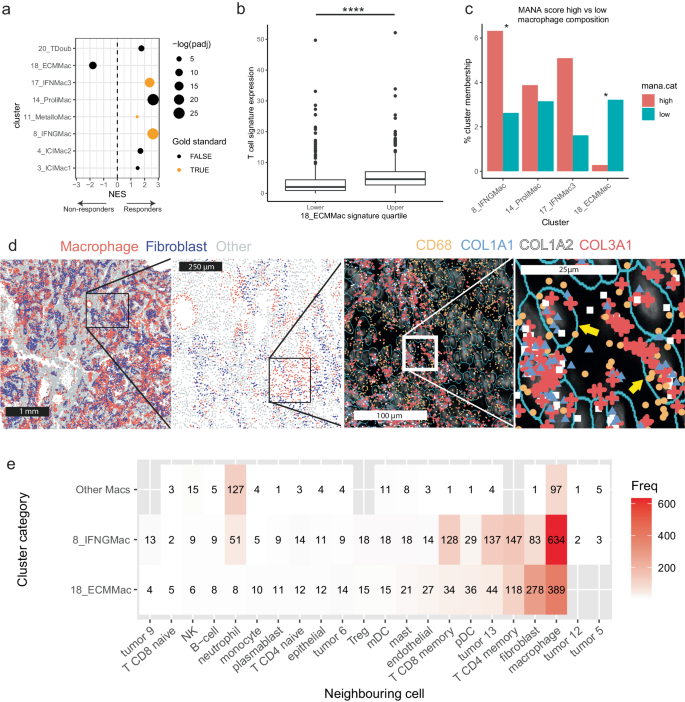
We examined the variations in expression of those signatures between responders and non-responders, while accounting for the impact of tumour kind, utilizing the CPI1000+ bulk RNAseq information together with DESeq295 and fast-gene set enrichment evaluation96 (strategies). We found a number of important relationships between our cluster signatures and response in each instructions. 20_TDoub and 8_IFNGMac signatures had been each considerably enriched in responding sufferers (fgsea, q-value = 0.001668273617609862 and 0.000000000013715289 respectively). This was anticipated, as the previous cluster consists of T cell doublets with macrophages, and essentially the most extremely upregulated gene within the latter was CXCL9, a chemokine identified to be concerned in T cell recruitment to tumours97. Equally, signatures 17_IFNMac3, 14_ProliMac, 11_MetalloMac, 4_ICIMac2 and 3_ICIMac1 had been considerably enriched in responders (fgsea, q-value = 0.000000017930358164, 0.000000000006084325, 0.036466593541472025, 0.002756081126333197 and 0.027749715367563134 respectively). Contrastingly, 18_ECMMac was considerably enriched in non-responding sufferers (fgsea, q-value = 0.000038213695118505; Fig. 4a). As this cluster is related to extracellular matrix modification, we hypothesized that the mechanism of resistance could also be as a result of T cell exclusion from the tumour. Nevertheless, in an evaluation of a normal T-cell signature (strategies) and its affiliation with the ECM signature, we noticed considerably larger (Two-sided Mann-Whitney U take a look at; ndecrease = 362, nhigher = 723; p < 0.0001; W = 74219; distinct samples) T-cell signatures related within the higher quartile of ECM signature samples within the CPI1000+ (Fig. 4b), indicating that normal T cell exclusion may not be the mechanism of affiliation between response and this cluster, and {that a} extra nuanced interplay between cells is perhaps at play. A lot of tumours in our TAM meeting evaluation had marked polarization in the direction of an 18_ECMMac state (Supplementary Be aware 1). By way of distribution by most cancers kind, most TAMs from 18_ECMMac had been from ccRCC (28.2%), adopted by HGSOC (15.4%) and CRC (14.9%).
a Dotplot exhibiting comparisons between responders and non-responders in expression of macrophage subset signatures in bulk expression information from the CPI1000+ cohort. Solely considerably totally different signatures between responders and non-responders after controlling for most cancers kind are proven. NES Normalized enrichment rating. “Gold-standard” signatures, which recognized their corresponding macrophage cluster in an all-celltype atlas with confidence (strategies), are indicated. b The generalised T cell signature is considerably larger within the higher quartile of 18_ECMMac signature within the CPI1000+ bulk RNA cohort in comparison with the decrease quartile. Two-sided Mann-Whitney U take a look at; n = 1084; ****p < 0.00000000000000022. Central line signifies median, field signifies interquartile vary, whiskers present 1.5x the IQR. c MANA rating evaluation exhibiting considerably totally different macrophage distributions in samples separated by MANA signature expression in T cells, comparability between higher and decrease quartiles of expression. The y-axis reveals the proportion of cells from the respective situation (MANA excessive or MANA low) belonging to the cluster indicated on the x-axis. Moderated two-sided T-test by way of Propeller87 with false-discovery fee correction for a number of testing; *q < 0.1. Precise q-values are 0.05962064 and 0.07801182 for clusters 8_IFNMac and 18_ECMMac respectively. d CosMx spatial information exhibiting a lung tumour wherein TAMs (expressing CD68), co-express a number of collagen transcripts. e Heatmap illustrating nearest-neighbour evaluation. ECM TAMs had been closest neighbours to different TAMs adopted by fibroblasts, while CXCL9-expressing TAMs had been closest neighbours with different TAMs adopted by T cells. Numbers point out the variety of cells in every class. MANA Mutation-associated neoantigen rating. Supply information are offered as a Supply Knowledge file.
One additional strand of investigation into the position of TAMs in ICI response is the interplay of TAMs and T cells. T cells exhibit adjustments to their transcriptional programme upon stimulation by cancer-associated neoantigens, together with upregulation of CXCL13 and MHC class II genes amongst others in lung most cancers98, which we collectively name the mutation-associated neoantigen rating, or MANA rating. We hypothesized that TAM-T cell interplay might play a task on this change, and compiled a second, smaller atlas of TAMs and T cells (see strategies) from lung cancers to evaluate whether or not there are adjustments in TAM cluster distribution in samples with T cells exhibiting totally different MANA scores. To attenuate the chance of kind I errors, we selected the highest 4 most vital signatures from the gene-set enrichment evaluation within the bulk CPI1000+ cohort above (14_ProliMac, 8_IFNGMac, 17_IFNMac3 and 18_ECMMac) and examined whether or not these clusters had considerably totally different proportions in lung tumour samples in higher vs decrease quartiles of MANA scores. We noticed considerably larger proportions of 18_ECMMac in samples within the decrease quartile of MANA scores in comparison with the higher quartile (Moderated two-sided T-test by way of Propeller with FDR correction for a number of testing, q = 0.07801182; Fig. 4c). Conversely, 8_IFNGMac was considerably enriched within the higher quartile of samples (Moderated two-sided T-test by way of Propeller with FDR correction for a number of testing, q = 0.05962064; Fig. 4c).
We subsequent sought to validate the presence of 18_ECMMac in tissue via using a spatial transcriptomic dataset comprising 5 tumours from NSCLC sufferers, generated on the NanoString CosMx platform. We calculated an 18_ECMMac signature rating on all cells with UCell, and noticed marked heterogeneity between samples, with one pattern containing numerous TAMs upregulating this signature, and others with only a few (Supplementary Fig. 8). Evaluation of the transcript expression revealed cells co-expressing CD68, COL1A1, COL1A2, and COL3A1 (Fig. 4d). We additionally recognized a number of putative fibroblasts, as recognized by comparability of mobile gene signatures to identified cell-type signatures99, that expressed CD68, doubtlessly indicative of an intermediate cell state in between fibroblasts and TAMs (Supplementary Fig. 9). We carried out a nearest neighbour evaluation, and located that the closest neighbouring cells to 18_ECMMac+ TAMs had been different TAMs adopted by fibroblasts (Fig. 4e), indicative of intercellular communication between these cell varieties. We additionally assessed the closest neighbours of 8_IFNGMac macrophages, discovering that the closest neighbours to 8_IFNGMac TAMs had been different TAMs, adopted by CD4 reminiscence T cells, most cancers cells and CD8 reminiscence T cells (Fig. 4e). In distinction, the closest neighbours to macrophages not belonging to both of those clusters had been TAMs, adopted by neutrophils and NK cells.
Our atlas augments current literature-based fashions, and types a complete reference for future research
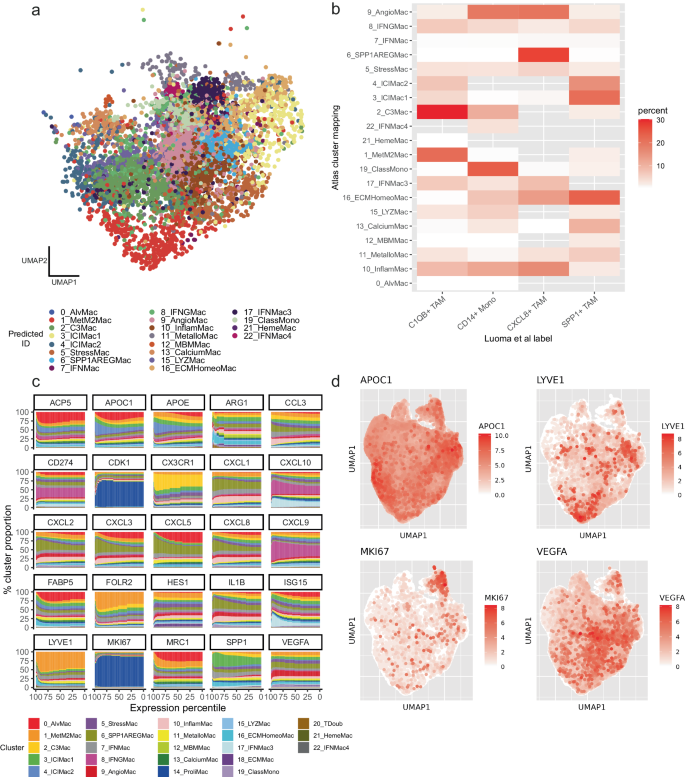
Along with facilitating a data-driven strategy to macrophage classification, the atlas additionally types a worthwhile useful resource for projection of novel datasets. We took a scRNAseq information from a current research on oral most cancers100 and projected the TAMs from this research onto the pan-cancer atlas (Fig. 5a). TAMs labeled as C1QB+ TAMs by the authors primarily mapped to our 2_C3Mac cluster (Fig. 5b), CD14+ Mono primarily mapped to 19_ClassMono cluster, CXCL8 + TAM mapped to 6_SPP1AREGMac and SPP1+ TAMs mapped to 16_ECMHomeoMac. Many TAMs additionally mapped to clusters apart from these most frequent mappings, resembling a proportion of the CD14+ Mono TAMs recognized by the authors mapping to our 9_AngioMac cluster, which was carefully associated to 19_ClassMono in our hierarchical clustering evaluation (Fig. 2c), maybe as a result of larger decision of our clustering within the atlas. There have been no 18_ECMMac TAMs detected within the oral most cancers dataset, indicating that this most cancers kind is negatively related to this pathway of macrophage differentiation. Markers extremely expressed within the unique clustering of the authors, resembling CXCL8 and SPP1, had been additionally extremely expressed within the mapped clusters. We additionally assessed the utility of the atlas in a novel spatial RNAseq dataset (Supplementary Be aware 2).
a UMAP projection of a novel oral most cancers dataset on to the atlas. b Heatmap exhibiting mapping of macrophage subsets within the oral most cancers dataset to subsets in our atlas. c Cluster distribution as a perform of magnitude of expression (percentile) of cells expressing subset-defining markers of Ma et al.14, indicating that some markers are cluster particular in our atlas, whereas others are broadly distributed throughout clusters. d UMAPs indicating cluster distribution of choose markers. Supply information are offered as a Supply Knowledge file.
Ideas on how greatest to characterize TAMs have more and more modified, shifting away from the M1/M2 dichotomy13 in the direction of extra difficult fashions encompassing macrophage stimuli and ontogeny14,15,101,102. Ma and colleagues suggest a seven-part mannequin that accounts for this variety, and likewise state that these classes lie on a spectrum, reflecting totally different levels of differentiation and stimuli of the macrophages. With the development of a pan-cancer macrophage atlas primarily based on high-resolution scRNAseq information, it’s doable to evaluate the dynamics of this spectrum and the markers that embody it. We took key marker genes outlined by Ma and colleagues and examined the cluster membership of the cells positively expressing these markers alongside every percentile of expression magnitude (Fig. 5c, d). This revealed various levels of heterogeneity in cluster membership, starting from markers principally dominated by one cluster, to markers that had been extra evenly unfold throughout all clusters, highlighting the pervasiveness of some markers within the macrophage panorama, and indicating that many markers are usually not indicative of macrophage differentiation state. Markers attributed to proliferating macrophages, together with MKI67 and CDK1 might be attributed to the previous class, the vast majority of cells expressing these genes belonging to cluster 14_ProliMac. Equally, cells expressing LYVE1 and FOLR2 primarily belonged to cluster 1_MetM2Mac, while cells expressing CXCL9 primarily belonged to cluster 8_IFNGMac. Markers distributed amongst numerous clusters included APOE, APOC1, ARG1 and HES1 (Fig. 5c).