TMIGD2 is extra extremely expressed on AML LSCs than on their regular counterparts
To discover whether or not the HHLA2-TMIGD2 pathway performs a novel position in regulating AML growth and mediating immune responses, we analyzed the cell floor expression of HHLA2 and TMIGD2 in 44 AML affected person samples by move cytometry (Fig. 1a and Supplementary Desk 1). Strikingly, TMIGD2, however not HHLA2, was selectively enriched in CD45dimSSClowLin–CD34+ cells (Fig. 1b, d). By analyzing a database containing AML samples with regular cells19, we discovered that the sample of HHLA2 and TMIGD2 protein expression was additionally in line with the information obtainable on the mRNA degree (Supplementary Fig. 1a, b). Moreover, outcomes from gene correlation evaluation indicated a optimistic affiliation between TMIGD2 and CD34 (Supplementary Fig. 1c). In The Most cancers Genome Atlas (TCGA) cohort, TMIGD2 was extra prominently expressed in FAB M0-M2 subtypes, acute promyelocytic leukemia (FAB M3), and FAB-M6 acute erythroid leukemia (Supplementary Fig. 1d)20. As LSCs primarily reside in CD34+CD38– and CD34+CD38+ compartments3,21, we additional examined these subsets of our AML specimens and located that TMIGD2 was extra extremely expressed on CD34+CD38– and CD34+CD38+ compartments compared with CD45dimSSClow unfractionated blasts (Fig. 1c). On condition that CD45RA, CD123, and IL1RAP have been beforehand recognized as LSC-specific markers8,11,22,23,24, we investigated their co-expression with TMIGD2 inside the CD34+ subpopulation. In 95% of 41 AML samples, AML cells from the CD34+TMIGD2+ subset expressed a minimum of one of many CD45RA, CD123 and IL1RAP markers (Supplementary Fig. 1e). Of notice, TMIGD2 was largely co-expressed with CD45RA inside each CD34+CD38– and CD34+CD38+ subpopulations (Supplementary Fig. 1f), suggesting that TMIGD2 is predominantly expressed on LSCs.
a Consultant move cytometry gating technique exhibiting HHLA2 and TMIGD2 expression on monocytes, stem cells (CD34+CD38–), progenitor cells (CD34+CD38+), and T cells (CD3+) of AML sufferers. Lin, CD14 and CD19. b Quantification of TMIGD2 expression on CD34+ and CD34– AML cells (n = 44). c Comparability of the expression degree of TMIGD2 in stem/progenitor subpopulations and bulk unfractionated blasts (n = 41). d Quantification of HHLA2 expression in AML CD34+ cells (n = 36) and regular CD34+ cells from CBU (n = 4) and NBM (n = 6) mononuclear cells. e Quantification of TMIGD2 expression in CD34+ AML cells (n = 40) relative to that in regular CD34+ cells (n = 10). Information are proven as proportion of optimistic (left) and imply fluorescence depth (MFI, proper) relative to the fluorescence minus one (FMO) management. f Comparability of TMIGD2 expression on CD3+ T cells from AML sufferers (n = 41) and wholesome donors (HD, n = 12). g Comparability of TMIGD2 expression on CD3+ T cells and CD34+ leukemia cells of human AML samples (n = 41). h Kaplan–Meier survival evaluation utilizing TCGA-AML dataset. The sufferers have been divided into two teams, excessive group (n = 79) > 25% of TMIGD2 degree, low group (n = 79) < 75% of TMIGD2 degree. The p worth was calculated by the log-rank take a look at. http://gepia2.cancer-pku.cn/#survival. Imply ± SD values are proven for Fig. 1d–f. Non-significant (ns), *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001 by two-tailed Scholar’s t take a look at (d–f) or paired Scholar’s t take a look at (b, c, g). Supply information are offered within the Supply Information file.
Apparently, we noticed an apparent upregulation of TMIGD2 in CD34+ AML cells in contrast with CD34+ HSPCs from 4 twine blood items (CBU) in addition to 6 regular grownup BM (NBM) samples (Fig. 1e and Supplementary Fig. 1g). Given the position of TMIGD2 in immune cells, we additionally examined TMIGD2 expression on T cells from AML specimens versus wholesome donors. As predicted, as a result of repetitive neoantigen stimulation, TMIGD2 expression on CD3+ T cells from AML samples was downregulated in contrast with wholesome donors (Fig. 1f). Furthermore, in people with AML, the expression of TMIGD2 was considerably larger in CD34+ AML cells than in CD3+ T cells (Fig. 1g). Kaplan–Meier evaluation revealed that the expression of TMIGD2 was a great predictor of AML development and thus indicative of a worse medical final result (Fig. 1h). Notably, AML sufferers harboring NPM1 mutation, who in lots of instances usually have <1% regular CD34+ cells11, expressed considerably decrease TMIGD2 (Supplementary Desk 2), suggesting that TMIGD2 is particularly expressed on the malignant CD34+ cells. Taken collectively, larger TMIGD2 expression in LSC-residing populations signifies that it might play a vital position in regulating the operate of LSCs and AML growth.
Lack of TMIGD2, however not HHLA2, inhibits AML growth and promotes myeloid differentiation
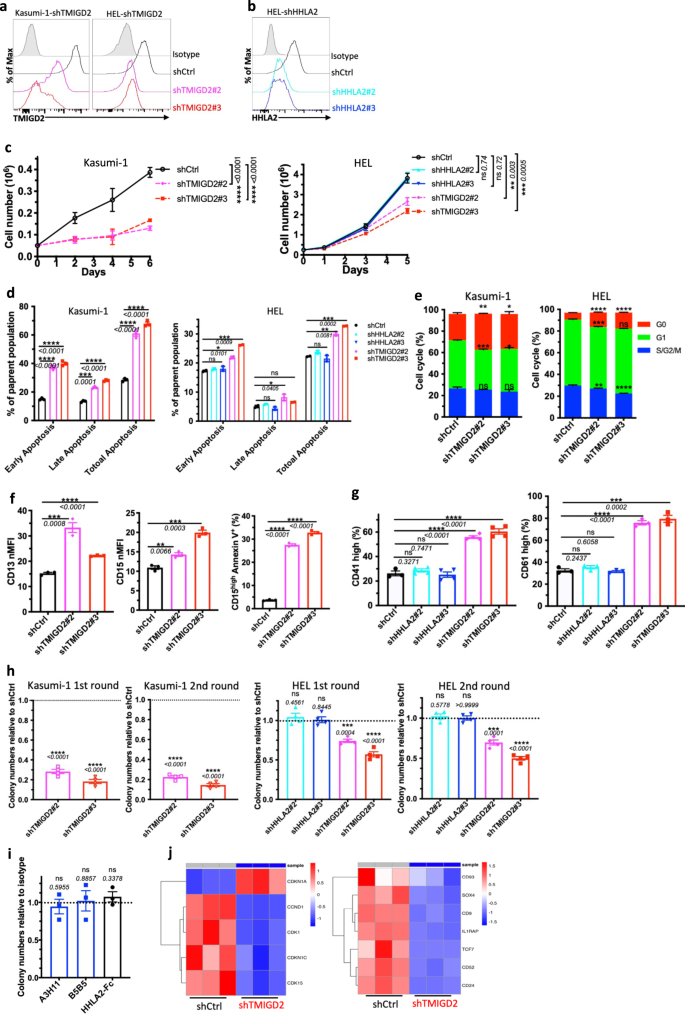
To analyze whether or not TMIGD2 performs a purposeful position in AML, we meant to determine human AML cell traces expressing TMIGD2. Utilizing the Most cancers Cell Line Encyclopedia (CCLE), we evaluated mRNA expression of TMIGD2 throughout >1000 totally different malignant cell traces, together with 39 AML cell traces25. TMIGD2 mRNA was extremely expressed in AML and T-cell acute lymphoblastic leukemia cell traces, whereas no different tumor varieties exhibited substantial TMIGD2 expression (Supplementary Fig. 2a). Human AML cell traces, together with HEL, K562, KG-1a, Kasumi-1 and ME-1, have been confirmed to specific cell floor TMIGD2 protein as analyzed by move cytometry (Supplementary Fig. 2b). To additional consider the features of TMIGD2 and HHLA2 in leukemia cells, we subsequent carried out small hairpin RNA (shRNA)-mediated knockdown of TMIGD2 (shTMIGD2) or HHLA2 (shHHLA2) in Kasumi-1, HEL and K562 cell traces (Fig. 2a, b and Supplementary Fig. 2c, d). Lack of TMIGD2, however not HHLA2, led to vital inhibition of leukemia cell development over the course of 6 days (Fig. 2c and Supplementary Fig. 2e). As well as, the dominant mobile phenotypes that resulted from TMIGD2 knockdown included a considerable induction of apoptosis (Fig. second and Supplementary Fig. 2f), a distinguished G0/G1 cell-cycle arrest (Fig. 2e, Supplementary Fig. 2g, h), a noticeable improve of differentiation (Fig. 2f, g and Supplementary Fig. 2i–ok) and a big lower of colony-forming cell (CFC) quantity (Fig. 2h, Supplementary Fig. 2l) in Kasumi-1, HEL and K562 AML cells. Nevertheless, these modifications weren’t noticed after HHLA2 knockdown (Fig. second, g, h and Supplementary Fig. 2f, ok). Additional evaluation by way of CFC assay revealed that self-renewal capability of HEL cells remained unchanged after remedy with anti-HHLA2 monoclonal antibodies (mAbs) or HHLA2-human Fc fusion protein (Fig. 2i), indicating that TMIGD2-mediated AML operate is HHLA2 unbiased. Notably, decreases in TMIGD2 expression, each on the floor protein and mRNA ranges, have been noticed when HEL cells have been handled with a differentiation-inducing agent, phorbol-12-myristate-13-acetate (PMA), suggesting that the downregulation of TMIGD2 is accompanied by myeloid differentiation (Supplementary Fig. 2m, n).
a Kasumi-1 (left) and HEL (proper) cells have been transduced with shRNA focusing on TMIGD2 and subjected to move cytometry evaluation. shCtrl, shRNA Management; shTMIGD2, TMIGD2 knockdown. Kasumi-1 is endogenously HHLA2 unfavourable. b Knockdown of HHLA2 in HEL cells. shHHLA2, HHLA2 knockdown. c Progress curves of Kasumi-1 (left) and HEL (proper) cells upon knockdown of HHLA2 or TMIGD2. d Quantification of apoptosis evaluation in Kasumi-1 (left) and HEL (proper) cells after HHLA2 or TMIGD2 knockdown. Early apoptosis, Annexin V+DAPI–; late apoptosis, Annexin V+DAPI+. e Quantification of cell-cycle evaluation in shCtrl and shTMIGD2 Kasumi-1 (left)/HEL (proper) cells. f Statistics of floor expression of differentiation markers CD13 (left) and CD15 (center) on shCtrl and shTMIGD2 Kasumi-1 cells. Proper, quantification of CD15excessiveAnnexin V+ cells in shCtrl and shTMIGD2 Kasumi-1 cells. g Statistics of floor expression of differentiation markers CD41 (left) and CD61 (proper) on shCtrl, shHHLA2 and shTMIGD2 HEL cells. h First spherical and second spherical colony-forming cell (CFC) counts of Kasumi-1 (left) and HEL (proper) cells after transduction with indicated lentiviruses. (500 cells/plate). i CFC counts of HEL cells after remedy with 50μg/ml anti-HHLA2 mAbs (A3H11 and B5B5) or HHLA2-hFc fusion protein. j Heatmap exhibiting differentially expressed genes associated to cell-cycle (left) and leukemia stemness (proper) in shCtrl versus shTMIGD2 HEL cells. Imply ± SEM values are proven for Fig. 2. ns, *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001 by two-tailed Scholar’s t take a look at. For c–i, colour dots signify technical replicates. Outcomes are consultant of three unbiased experiments. Supply information are offered within the Supply Information file.
To analyze the impact of TMIGD2 on transcriptional features in AML cells, we carried out gene expression analyses by RNA-sequencing (RNA-seq) in shCtrl and shTMIGD2 HEL cells (Supplementary Fig. 2o). Gene set enrichment evaluation (GSEA) revealed that pathways enriched amongst differentially expressed genes included mitotic spindle, interferon alpha response, and Runx1 regulated transcription of genes concerned in differentiation of HSCs (Supplementary Fig. 2p). Theses pathways mirrored differentiation and cell biking as potential phenotypes related to TMIGD2 knockdown in HEL cells. Among the many genes with vital modifications upon TMIGD2 depletion have been components intently related to cell-cycle (CDKN1A and CCND1) and leukemia stemness (CD93 and IL1RAP; Fig. 2j).
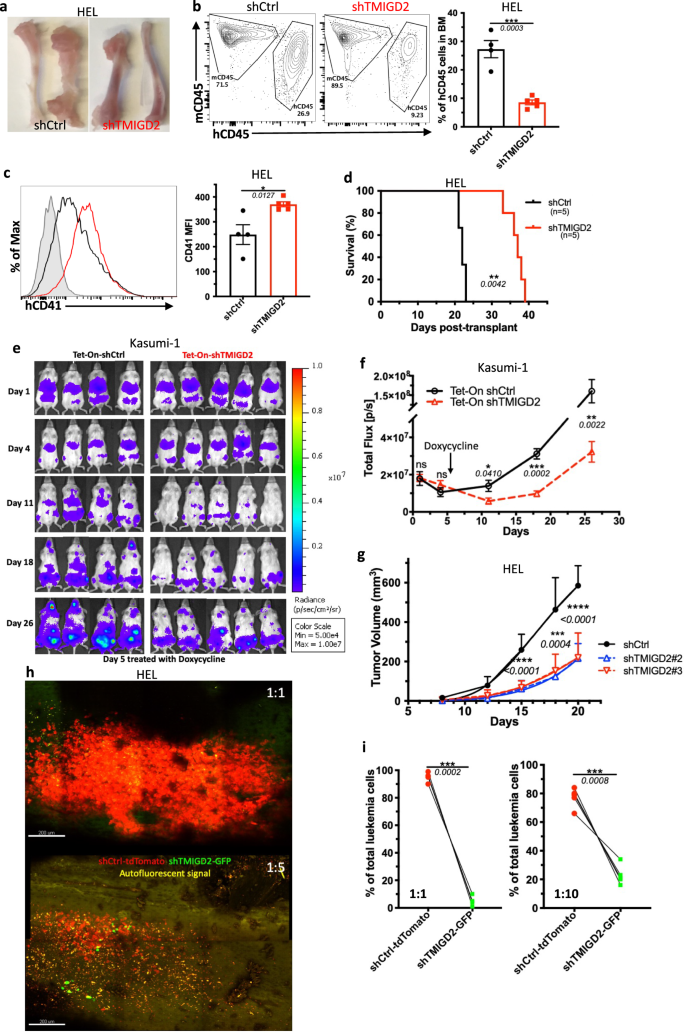
To evaluate the position of TMIGD2 in leukemogenesis in vivo, we firstly established an AML cell line-derived xenograft mannequin by intravenously transplanting the shCtrl or shTMIGD2 AML cells into sublethally irradiated NSG mice. As proven in Fig. 3a–d and Supplementary Fig. 3a, b, we noticed that NSG-recipients of shTMIGD2 HEL and shTMIGD2 K562 cells developed and died of AML considerably slower than did recipients of shCtrl HEL and shCtrl K562 cells. The delayed growth of AML within the absence of TMIGD2 was related to a decrease proportion of leukemic cells within the BM and spleen (Fig. 3a, b and Supplementary Fig. 3a), in addition to an induction of differentiation (Fig. 3c). Subsequent, we implanted NSG mice with Tet-On shCtrl or Tet-On shTMIGD2 Kasumi-1 cells and handled the NSG-recipients with doxycycline that may induce Tet-On shRNA to silence TMIGD2 (Supplementary Fig. 3c). When induced at day 5 after cell transplantation, the knockdown of TMIGD2 slowed AML growth (Fig. 3e, f). Furthermore, depletion of TMIGD2 additionally considerably arrested tumor development in HEL and K562 subcutaneous xenograft fashions of AML in NSG mice (Fig. 3g and Supplementary Fig. 3d, e). General, these information strongly implicate the significance of TMIGD2 for AML growth and upkeep.
a Consultant photos of tibia and femur from mice engrafted with shCtrl or shTMIGD2 HEL cells. b Consultant move cytometry plots (left) and statistics (proper) of BM cells from mice inoculated with shCtrl or shTMIGD2 HEL cells. c Consultant move cytometry histogram (left) and quantification (proper) of CD41 expression on BM hCD45+ cells remoted from NSG mice engrafted with shCtrl or shTMIGD2 HEL cells. d Kaplan–Meier survival curves of mice transplanted with shCtrl or shTMIGD2 HEL cells (n = 5 per group, three unbiased experiments). The p worth was calculated by the log-rank take a look at. e Photographs of NGS mice that have been engrafted with Tet-On shRNAs Kasumi-1 cells and handled with doxycycline at day 5 put up engraftment to induce shRNA expression. Radiance (p/sec/cm2/Sr). f Leukemia development was quantified by bioluminescence. Complete flux [p/s] for Tet-On shCtrl versus Tet-On shTMIGD2 Kasumi-1 cell line-derived xenograft mice. g Common development curves of subcutaneous shCtrl and shTMIGD2 HEL tumors in NSG mice. h Consultant intravital 3D flattened photos of shCtrl-tdTomato and shTMIGD2-GFP HEL leukemia cells within the BM of NSG-recipients. High, cell ratio for i.v. injection shCtrl-tdTomato: shTMIGD2-GFP = 1:1; backside, cell ratio for i.v. injection shCtrl-tdTomato: shTMIGD2-GFP = 1:5. i Comparability of shCtrl-tdTomato and shTMIGD2-GFP cell proportion within the BM of NSG-recipients engrafted with combined shCtrl-tdTomato and shTMIGD2-GFP HEL cells on the ratio of 1:1 (left) or 1:10 (proper). Imply ± SEM values are proven for Fig. 3. *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001 by two-tailed Scholar’s t take a look at (b, c, f, g) or paired Scholar’s t take a look at (i). For b–c and i, colour dots signify particular person mice. Outcomes are consultant of three unbiased experiments. Supply information are offered within the Supply Information file.
We subsequent used intravital two-photon microscopy to check the behavioral response of AML cells within the BM upon TMIGD2 depletion. By intravenous injection of combined cells at totally different ratios, we discovered that shCtrl-tdTomato HEL cells turned the predominant cells inside the tibia BM (Fig. 3h, i), suggesting that TMIGD2 is crucial for the enlargement of leukemic cells within the BM. Moreover, time-lapse movies of NSG-recipients injected concomitantly with shCtrl-tdTomato and shTMIGD2-GFP HEL cells revealed that leukemic cell motility was considerably decreased after TMIGD2 knockdown (Supplementary Fig. 3f), and movies additionally confirmed extra shCtrl-tdTomato cells recirculating inside blood sinusoid areas (Supplementary Fig. 3g). To substantiate these outcomes, leukemic cells inside the BM vascular area of interest of recipient NSG mice have been flash-labeled by intravenous administration of fluorophore-conjugated antibody26. As anticipated, the next frequency of labeled shCtrl-tdTomato leukemic cells have been noticed (Supplementary Fig. 3h). These information, along with the presence of extra shCtrl than shTMIGD2 leukemic cells within the spleen of NSG-recipients (Supplementary Fig. 3a), point out that TMIGD2 contributes to dissemination of leukemic cells.
TMIGD2+ AML cells are enriched in LSC exercise
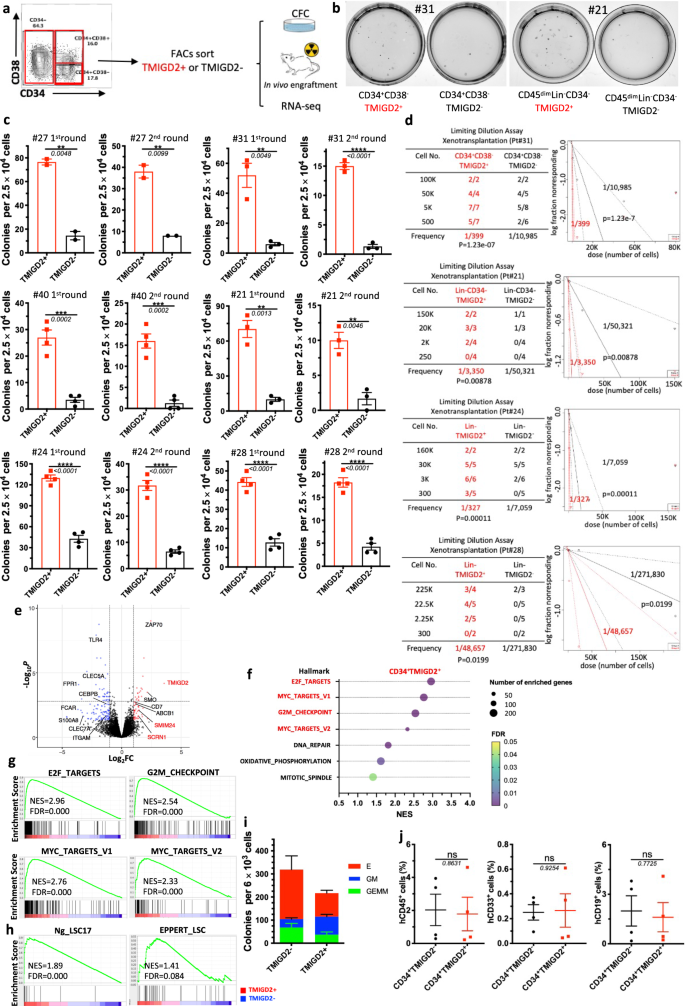
In most human AML, LSCs reside predominantly inside the CD34+ subpopulation of AML blasts, amongst which they’re extremely enriched based mostly on CD38 negativity and expression of different beforehand recognized floor markers3,6,12. Contemplating that TMIGD2 was often expressed in CD34+CD38– and CD34+CD38+ cell fractions, we hypothesized that the expression of TMIGD2 may outline subset of leukemic cells with distinct leukemogenesis properties. To check this speculation, we used fluorescence-activated cell sorting (FACS) to fractionate major human AML specimens into CD34+CD38–TMIGD2+ and CD34+CD38–TMIGD2– subpopulations, or CD34+CD38+TMIGD2+ and CD34+CD38+TMIGD2– subpopulations in samples that CD34+CD38– subset was absent (Supplementary Desk 3). CFC assays have been then carried out (Fig. 4a). Clonogenic means was dramatically larger within the TMIGD2+ fraction in contrast with the TMIGD2– fraction of CD34+CD38–/CD34+CD38+ cells upon serial replating, suggesting a discount in LSC frequency and self-renewal in CD34+CD38– (or CD34+CD38+) TMIGD2– subpopulation (Fig. 4b, c and Supplementary Fig. 4b). Since we noticed TMIGD2 expression in CD34– cells from a small subset of AML sufferers whose CD34 expression was restricted, we investigated whether or not TMIGD2 may enrich for purposeful LSCs in CD34– AML. CD34–TMIGD2+ AML cells confirmed enhanced in vitro clonogenicity compared with their TMIGD2– counterpart (Fig. 4b, c). Comparable outcomes have been obtained when CFC assays have been performed utilizing FACS-purified TMIGD2+ and TMIGD2− blasts no matter CD34 and CD38 expression (Fig. 4c and Supplementary Fig. 4a). To instantly enumerate the frequency of LSCs, a limiting dilution xenotransplantation experiment was carried out with FACS-purified subsets27. In keeping with the outcomes of CFC assay, the CD34+CD38–TMIGD2+, CD34–TMIGD2+ and Lin–TMIGD2+ subpopulations had a considerably larger LSC frequency than corresponding TMIGD2– subpopulations, with enhanced means to repopulate NSG mice in xenotransplantation assays (Fig. 4d, Supplementary Fig. 4c–e, and Supplementary Desk 3).
a Schematic for move cytometry sorting and subsequent CFC assay and in vivo xenotransplantation. The very best (TMIGD2+) and lowest (TMIGD2–) TMIGD2-expressing blasts (high and backside 15%, respectively) from the CD34+CD38–, CD34+CD38+ and CD34− AML specimens have been FACS-purified and used for CFC assays, xenotransplantation and RNA-seq evaluation. b Consultant picture of CFC plates seeded with FACS-purified TMIGD2+ and corresponding TMIGD2– AML affected person cells with respect to colony formation after 12 days tradition. Left, Pt#31; proper, Pt#21. c First spherical (left) and second spherical (proper) CFC assays utilizing FACS-purified TMIGD2+ or TMIGD2– cells of CD34+CD38− (Pt#27 and Pt#31), CD34+CD38+ (Pt#40), CD34– (Pt#21) and Lin– (Pt#24 and Pt#28) subpopulations from particular person AML sufferers. d Limiting dilution assay. Desk (left) exhibiting totally different numbers of FACS-purified cells and recipient NSG mice used for the xenotransplantation. Graph (proper) exhibiting the frequency of LSC in TMIGD2+ and TMIGD2− subpopulations. The limiting dilution evaluation was carried out utilizing ELDA (Excessive Limiting Dilution Evaluation) software program. e The volcano plot exhibiting differentially expressed genes from RNA-seq evaluation on CD34+TMIGD2+ and CD34+TMIGD2– cells sorted from six major AML affected person samples. f Scattergrams of the highest pathways that have been considerably enriched in CD34+TMIGD2+ cells based mostly on GSEA. g GSEA plots exhibiting enrichment of gene units for E2F targets, Myc targets and G2M checkpoints in CD34+TMIGD2+ versus CD34+TMIGD2– teams from six major AML affected person samples. h GSEA plots exhibiting the stemness signatures have been enriched in CD34+TMIGD2+ cells. i CFC assay was carried out with FACS-purified TMIGD2+ or TMIGD2– cells from regular CD34+ HSPCs. j Percentages of hCD45+, hCD33+ and hCD19+ cells within the BM of NSG-recipient mice at 3 months put up transplantation of FACS-purified TMIGD2+ or TMIGD2– cells from CD34+ HSPCs. Imply ± SEM values are proven for Fig. 4c, i, j. ns by two-tailed Scholar’s t take a look at. Normalized enrichment rating (NES) and false discovery price (FDR) are proven for Fig. 4g, h. Coloration dots in c and i signify technical replicates. Coloration dots in j signify particular person mice. Outcomes are consultant of three unbiased experiments. Supply information are offered within the Supply Information file.
To comprehensibly perceive the genes and pathways related to TMIGD2 expression in LSCs, transcriptome-wide RNA-seq was performed on FACS-purified CD34+TMIGD2+ and CD34+TMIGD2– subfractions of six major AML specimens. The RNA-seq evaluation revealed {that a} whole of 453 genes have been considerably differentially expressed by a minimum of 2-fold (FDR < 0.05; Fig. 4e). Amongst these differentially expressed genes, the CD34+TMIGD2+ leukemic cells had elevated expression of a number of genes classically related to LSC signatures and poor prognosis, reminiscent of SCRN1, SMIM24, and ABCB1. In distinction, CD34+TMIGD2– leukemic cells exhibited larger expression of monocyte/granulocyte-associated genes (e.g., ITGAM, CLEC5A, FPR1, CEBPB, and S100A8; Fig. 4e). Importantly, GSEA confirmed that the highest seven pathways enriched in CD34+TMIGD2+ fraction consisted of E2F targets, MYC targets, and G2M checkpoints (Fig. 4f, g), which was in line with our findings that CD34+TMIGD2+ cells generated extra colonies and induced leukemia rather more effectively than TMIGD2− counterparts (Fig. 4b–d). Against this, we discovered vital enrichment of a set of functionally necessary signaling pathways in CD34+TMIGD2– subpopulation, together with TNF-α signaling, inflammatory response, apoptosis, and the p53 pathway (Supplementary Fig. 4f, g). Furthermore, CD34+TMIGD2+ subpopulation was related to the established 17-gene stemness (LSC17) and LSC signatures (Fig. 4h), whereas corresponding TMIGD2– fraction was correlated with myeloid cell growth, hematopoiesis maturation, and downregulation of HOXA9 and MEIS1 targets (Supplementary Fig. 4g).
To analyze the self-renewal and differentiation potential of TMIGD2+ and TMIGD2– subsets in regular HSPCs, a CFC assay and xenotransplantation have been carried out utilizing FACS-purified CD34+TMIGD2+ and CD34+TMIGD2– cells from CBU and NBM samples. Each fractions have been discovered to kind GEMM (Granulocyte/Erythrocyte/Macrophage/Megakaryocyte), GM (granulocyte/macrophage), and E (erythroid) colonies (Fig. 4i). Nevertheless, most GM grew out of the CD34+TMIGD2+ fraction, whereas E originated from the CD34+TMIGD2– fraction (Fig. 4i and Supplementary Fig. 4h, i). In vivo HSPCs exercise was indicated by the presence of each hCD45+CD19+ lymphoid and hCD45+CD33+ myeloid cells within the BM of NSG mice 12 weeks after transplantation (Supplementary Fig. 4j). When transplanted at equal cell doses, each lymphoid and myeloid engraftments have been detected with an identical frequency in NSG mice engrafted with both TMIGD2+ or TMIGD2– subsets (Fig. 4j). Taken collectively, these outcomes point out that, though regular purposeful HSPCs reside in each CD34+TMIGD2+ and CD34+TMIGD2− compartments, LSCs reside predominantly inside the CD34+TMIGD2+ compartment in AML.
TMIGD2 is crucial for sustaining major human AML LSC functionality
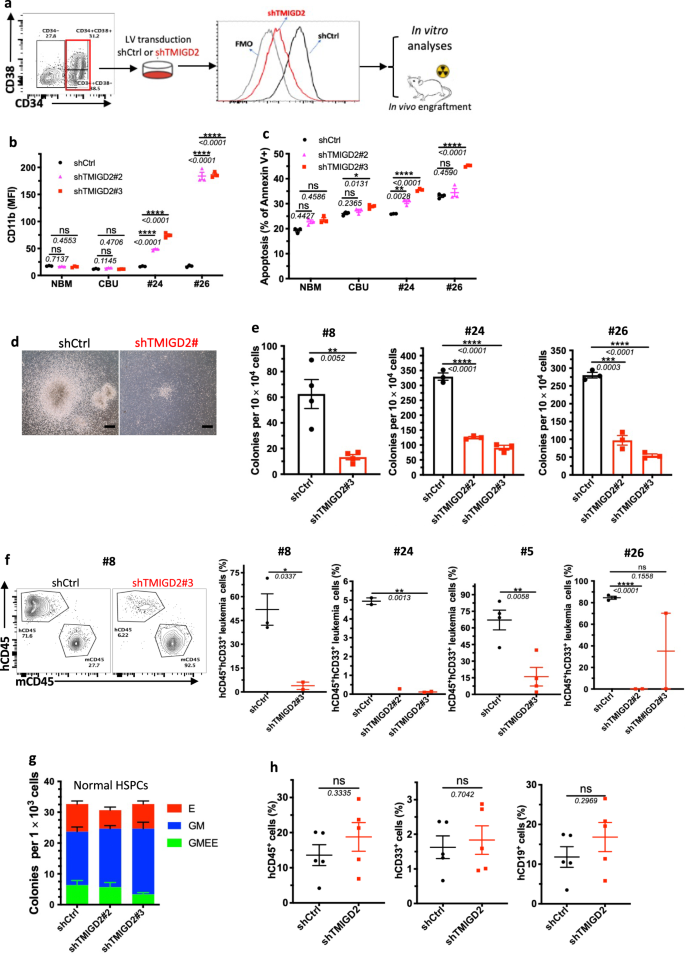
To outline the direct position of TMIGD2 in sustaining LSC operate and leukemogenesis, FACS-purified CD34+CD38– or CD34+CD38+ major AML cells from sufferers have been transduced with lentivirus expressing shCtrl or shTMIGD2. We subsequent performed in vitro analyses and xenotransplantation experiments utilizing the shCtrl and shTMIGD2 major AML cells (Fig. 5a). TMIGD2 knockdown resulted in elevated expression of differentiation marker CD11b and apoptotic marker annexin V in major AML cells however not in NBM and CBU (Fig. 5b, c and Supplementary Fig. 5a), indicating that TMIGD2 is particularly necessary for LSCs. As well as, we discovered that TMIGD2 depletion considerably decreased the scale and variety of colonies derived from major AML cells (Fig. 5d, e), supporting a job for TMIGD2 in selling proliferation and clonogenic potential of leukemic cells. In settlement with the in vitro information, knockdown of TMIGD2 resulted in considerably delayed human AML development (Pt#5 and Pt#8), lowered engraftment potential (Pt#24 and Pt#26), and extended survival (Pt#5) in AML PDX fashions (Fig. 5f and Supplementary Fig. 5b, c), indicating that TMIGD2 is crucial for the event of AML.
a Schematic for move cytometry sorting technique, lentivirus transduction and subsequent experimental design of in vitro and in vivo assays. b Expression of myeloid lineage marker CD11b after knockdown of TMIGD2 in major CD34+ cells remoted from NBM, CBU and AML affected person samples (Pt#24 and Pt#26). c Share of Annexin V+ cells in shTMIGD2 in contrast with shCtrl major CD34+ cells as assessed by move cytometry. d, e Consultant morphology (d, Pt#8) and quantification (e) of colonies derived from sorted major CD34+ AML cells transduced with lentiviruses focusing on shCtrl or shTMIGD2. Scale bar: 100 μm for colonies. f Consultant move cytometry plots (left) and engraftment potential (proper) of PDX mouse fashions established with major AML cells that have been transduced with lentiviruses focusing on shCtrl or shTMIGD2. g CFC assay utilizing regular CD34+ HSPCs upon knockdown of TMIGD2 (n = 3). h Percentages of hCD45+, hCD33+ and hCD19+ cells within the BM of NSG-recipient mice at 3 months put up transplantation of shCtrl or shTMIGD2 CD34+ HSPCs. Imply ± SEM values are proven for Fig. 5. ns, *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001 by two-tailed Scholar’s t take a look at. Coloration dots in b, c, e, g signify technical replicates. Coloration dots in f and h signify particular person mice. Outcomes are consultant of three unbiased experiments. Supply information are offered within the Supply Information file.
Subsequent, we explored whether or not TMIGD2 is required for regular hematopoiesis. We knocked down TMIGD2 by shRNAs in human-derived-CD34+ cells after which carried out CFC assay and xenotransplantation. As proven in Fig. 5g, the comparable colony-forming means was noticed through the use of shCtrl or shTMIGD2 CD34+ cells. In contrast with NSG mice engrafted with shCtrl CD34+ cells, mice engrafted with shTMIGD2 CD34+ cells exhibited comparable percentages of whole hCD45+ cells, hCD19+ lymphoid cells and hCD33+ myeloid cells within the BM (Fig. 5h), suggesting that TMIGD2 deletion has no detectable affect on regular human CD34+ HSPCs.
TMIGD2 regulates the operate of LSC by the ERK1/2-p90RSK-CREB pathway
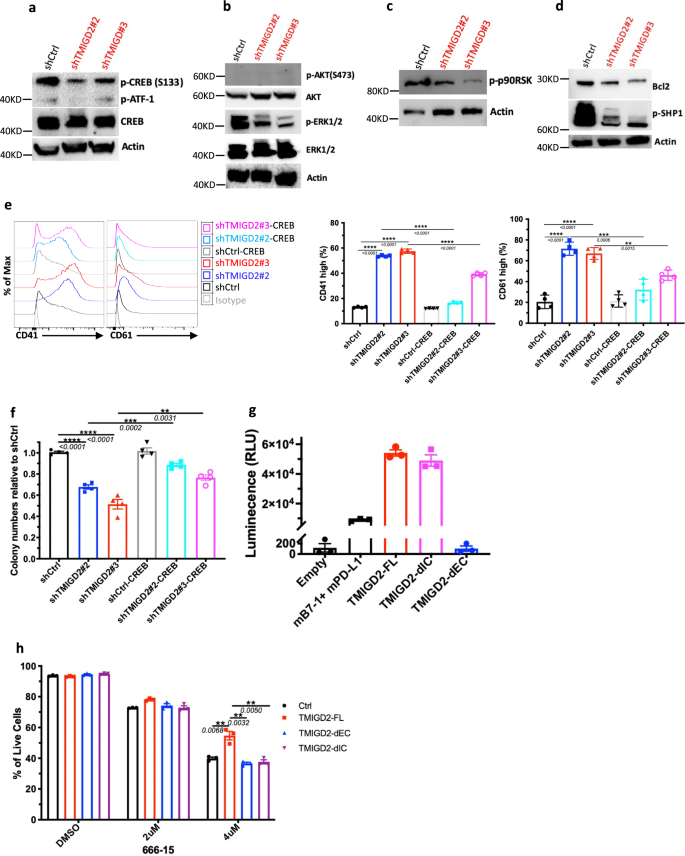
We subsequent sought to know how TMIGD2 depletion influences signaling pathways in AML cells. Utilizing a human phospho-kinase array, we first in contrast signaling protein ranges in shCtrl and shTMIGD2 HEL cells. We discovered that the abundance of phosphorylated CREB (p-CREB) was considerably lowered upon TMIGD2 knockdown (Supplementary Fig. 6a), which was verified by Western blot (Fig. 6a). Contemplating that CREB can act as both a direct or an oblique substrate for ERK and AKT pathways, which play vital roles within the transmission of proliferative indicators from membrane sure receptors in AML cells28, we subsequent examined whether or not depletion of TMIGD2 may have an effect on these pathways. Western blot evaluation confirmed that p-ERK1/2 however not p-AKT protein ranges have been decreased in shTMIGD2 HEL cells in comparison with management (Fig. 6b). In settlement with these remark in HEL cells, knockdown of TMIGD2 decreased degree of p-CREB and p-ERK1/2 in major CD34+ AML cells however not in CD34+ cells remoted from NBM and CBU (Supplementary Fig. 6b). Moreover, it has been proven that ribosomal S6 kinases (RSKs) phosphorylate CREB, selling myeloid cell proliferation and survival by induced expression of Bcl-2 and cyclin-A29,30. The Western blot analyses confirmed that lack of TMIGD2 in AML cells led to an apparent lower of p-p90RSK and Bcl-2 (Fig. 6c, d). Moreover, a decrease p-SHP-1 degree was noticed in TMIGD2 knockdown cells compared with management, suggesting that SHP-1 is likely to be partially concerned in TMIGD2-mediated sign transduction pathways in AML (Fig. 6d). To additional affirm that CREB phosphorylation is required for the phenotypes seen in TMIGD2-depleted leukemic cells, we carried out rescue experiments by introducing retrovirus encoding wild sort CREB into shCtrl and shTMIGD2 HEL cells (Supplementary Fig. 6c). The ectopic expression of CREB was able to considerably rescuing each the differentiation and colony-forming means ensuing from endogenous TMIGD2 depletion in HEL cells (Fig. 6e, f). General, these information recommend that the ERK1/2-p90RSK-CREB pathway functionally contributes to the results of TMIGD2 in AML.
a Western blot evaluation of phospho-CREB (p-CREB) and whole CREB degree modifications in HEL cells upon TMIGD2 knockdown. b–d Western blot photos exhibiting the extent modifications of p-ERK1/2 (b), p-AKT (b), p-p90RSK (c), Bcl2 (d), and p-SHP1 (d) in shCtrl versus shTMIGD2 HEL cells. e Consultant move cytometry histograms (left) and quantification (proper) of CD41 and CD61 expression on HEL cells engineered to specific CREB with or with out TMIGD2 knockdown. f Colony-forming means of HEL cells from Fig. 6e (500 cells/plate). g NanoBit proximity assay exhibiting TMIGD2 binds with TMIGD2 on the identical cell floor in a cis interplay. Mouse PD-L1 (mPD-L1) and mB7-1 kind a heterodimer, which was served as a optimistic management. TMIGD2-FL, full size TMIGD2. TMIGD2-dEC, a mutant TMIGD2 with deleted extracellular area. TMIGD2-dIC, a mutant TMIGD2 with deleted intracellular area. h Impact of a CREB inhibitor 666-15 on apoptosis of THP-1 cells overexpressing vector Ctrl, TMIGD2-FL, TMIGD2-dEC and TMIGD2-dIC. Imply ± SEM values are proven for Fig. 6. **p < 0.01, ***p < 0.001, and ****p < 0.0001 by two-tailed Scholar’s t take a look at. Coloration dots in e–h signify technical replicates. Outcomes are consultant of three unbiased experiments. Supply information are offered within the Supply Information file.
Because the position of TMIGD2 in AML is unbiased of engagement with its identified ligand HHLA2 (Figs. 1d, 2d, g, h, and 7e, and Supplementary Fig. 7f), we hypothesized that TMIGD2 signaling in AML is triggered by: (1) a brand new binding associate; (2) a gain-of-function mutation; (3) trans- and/or cis-homophilic dimerization. To check these hypotheses, we first carried out cell-based high-throughput screening for a possible new binding associate of TMIGD2 (Supplementary Fig. 6d). Amongst 5477 human plasma membrane proteins and cell floor tethered human secreted proteins, in addition to 371 heterodimers, no new interactor, along with HHLA2, was recognized (Supplementary Fig. 6d). Subsequent, to research the genetic components accountable for the activation of TMIGD2 in AML, TMIGD2 cDNA of three AML specimens (Pt#19, #20 and #26) and HEL cell line have been sequenced. In settlement with cBioPortal database (Supplementary Fig. 6e)31, cDNA sequencing of cell line and first samples confirmed restricted alterations inside intracellular area (IC) and transmembrane area (TM) of TMIGD2 (Supplementary Fig. 6f), suggesting {that a} gain-of-function mutation is unlikely the reason for TMIGD2 signaling in AML. We subsequent decide whether or not TMIGD2 binds with TMIGD2 in a trans method by staining HHLA2/3T3 or TMIGD2/3T3 cells with TMIGD2 mouse Fc fusion protein adopted by secondary antibody staining. TMIGD2 was noticed to bind with HHLA2, however not TMIGD2 (Supplementary Fig. 6g). To check whether or not TMIGD2 types dimers on the identical cell floor, we carried out the NanoBit proximity assay by co-transfection of constructs comprising proteins of curiosity fused to small-bit (SmBit) and large-bit (LgBit) (Supplementary Fig. 6h). TMIGD2-full size (FL) and TMIGD2-deleted IC (dIC), however not TMIGD2-deleted extracellular area (dEC), generated luminescent sign, demonstrating that EC of TMIGD2 is required for cis homodimer formation (Fig. 6g).
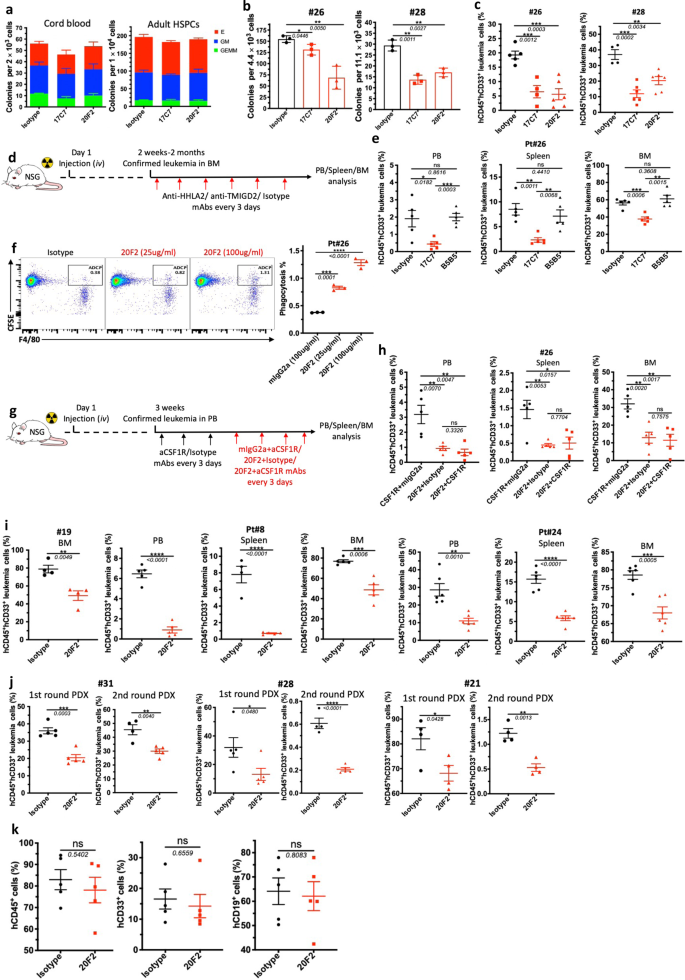
a CFC assay utilizing CBU (left) and NBM (proper) -derived CD34+ HSPCs handled with isotype management or anti-TMIGD2 mAbs (17C7 and 20F2). b CFC counts of major CD34+ AML cells after remedy with isotype management or anti-TMIGD2 mAbs (17C7 and 20F2). c Share of hCD45+CD33+ leukemia cells in PB (Pt#26) and BM (Pt#28) of AML PDX mouse fashions after remedy with isotype management or anti-TMIGD2 mAbs (17C7 and 20F2). d Schematic for engraftment of major AML specimens and in vivo remedy with anti-HHLA2, anti-TMIGD2 and isotype mAbs. e Frequency of hCD45+CD33+ leukemia cells in PB, spleen and BM after remedy with mIgG1 isotype management, anti-HHLA2 (B5B5, mIgG1) and anti-TMIGD2 (17C7, mIgG1) mAbs. f Phagocytosis of major AML cells by mouse BM derived macrophages within the presence of 20F2 or mIgG2a isotype management. g Experimental schematic of in vivo macrophage depletion with anti-CSF1R remedy (black arrow), adopted by 20F2 and anti-CSF1R mAbs mixture remedy (crimson arrow). aCSF1R, anti-CSF1R. h Share of hCD45+CD33+ leukemia cells in PB, spleen and BM in Fig. 7g. i Share of hCD45+CD33+ leukemia cells in PB, spleen and BM of AML PDX mouse fashions established with R/R AML samples after remedy with isotype management or 20F2. j Frequency of hCD45+CD33+ leukemia cells within the BM of the primary spherical (left) and second spherical (proper) PDX mouse fashions. The first PDX mice have been handled with isotype management versus 20F2. The second spherical NSG-recipients acquired no additional 20F2 remedy. ok Share of human CD45+, CD33+ and CD19+ cells within the BM of NSG-recipients engrafted with regular HSPCs remoted kind twine blood after remedy with mIgG2a isotype management or 20F2. Imply ± SEM values are proven for Fig. 7. ns, *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001 by two-tailed Scholar’s t take a look at. Coloration dots in a, b, and f signify technical replicates. Coloration dots in c, e, h and i–ok signify particular person mice. Outcomes are consultant of three unbiased experiments. Supply information are offered within the Supply Information file.
To know whether or not the EC and IC domains of TMIGD2 are vital for the regulation of CREB in AML cells, TMIGD2-FL, TMIGD2-dEC, and TMIGD2-dIC have been expressed in THP-1 cells adopted by remedy with a CREB inhibitor 666-1532. THP-1 cells overexpressing TMIGD2-FL have been extra immune to 666-15-induced apoptosis than cells overexpressing both TMIGD2-dEC or TMIGD2-dIC (Fig. 6h and Supplementary Fig. 6i), indicating that each EC and IC domains of TMIGD2 are necessary for its operate in AML. We then launched mutations at amino acids tyrosine 192 (Y192F), serine 220 (S220A), and tyrosine 222 (Y222F) inside the cytoplasmic tail of TMIGD2. THP-1 cells harboring TMIGD2-Y192F and TMIGD2-S220A confirmed larger proportion of reside cells than TMIGD2-dIC and TMIGD2-Y222F after 666-15 remedy, suggesting that tyrosine 222 is concerned in TMIGD2-mediated downstream signaling pathways in AML (Supplementary Fig. 6j).
Monoclonal antibodies in opposition to TMIGD2 show anti-leukemia exercise
The event of potent and selective TMIGD2 mAbs make it doable to guage the therapeutic advantages of focusing on TMIGD2 in AML. To this finish, we generated anti-TMIGD2 mAbs recognizing distinct epitopes of TMIGD2, together with clones 17C7 (mouse IgG1, mIgG1) and 20F2 (mIgG2a), which have been in a position to disrupt the TMIGD2 cis-homodimerization and fully block and partially block the interplay between HHLA2 and TMIGD2, respectively (Supplementary Fig. 7a–c). When handled with 17C7 and 20F2, TMIGD2+ major AML samples confirmed vital lower in colony-forming means; nevertheless, regular HSPCs from CBU and grownup BM, in addition to TMIGD2– major AML cells exhibited no vital distinction (Fig. 7a, b and Supplementary Fig. 7d), suggesting that anti-TMIGD2 mAbs selectively inhibit TMIGD2+ major AML cells. We subsequent assessed the therapeutic efficacy of 17C7 and 20F2 in vivo utilizing clinically related AML PDX fashions. The anti-leukemic results of 17C7 and 20F2 have been confirmed by the discount of human CD45+CD33+ leukemia cells in peripheral blood (PB) and BM of NSG-recipients following remedies (Fig. 7c and Supplementary Fig. 7e). Notably, each 17C7 and 20F2 effectively lowered leukemia burden in PDX mannequin engrafted with pt#28, who had progressed after a number of earlier remedies, together with Bcl-2 inhibitor venetoclax (Fig. 7c).
By taking the benefit that each of our anti-TMIGD2 mAb 17C7 and anti-HHLA2 mAb B5B5 can fully block the interplay between HHLA2 and TMIGD2, we handled AML PDX fashions with both 17C7 or B5B5 to rule out the likelihood that HHLA2-TMIGD2 axis contributes to the event of AML in vivo (Fig. 7d). As anticipated, 17C7 however not B5B5 remedy considerably lowered leukemia cells within the PB, spleen, and BM (Fig. 7e). Furthermore, HHLA2 expression was undetectable in leukemia cells from BM of each mouse teams handled with 17C7 and B5B5 (Supplementary Fig. 7f). These information additional affirm that the operate of TMIGD2 in AML shouldn’t be as a result of HHLA2-TMIGD2 interplay.
As 20F2 is a mouse IgG2a mAb, we subsequent explored whether or not its anti-leukemia results might be partially attributed to antibody-dependent mobile phagocytosis (ADCP) by performing a BM-derived macrophage coculture system. 20F2 opsonized major leukemia cells resulted in a considerably larger ADCP exercise in contrast with management mIgG2a (Fig. 7f), suggesting that 20F2 could each block oncogenic TMIGD2 signaling and induce ADCP. To additional analyze whether or not the direct involvement of macrophages that elicit ADCP in vivo is critical for 20F2 mAb therapeutic efficacy, we handled PDX NSG mice with anti-CSF1R mAb to deplete macrophages (Fig. 7g and Supplementary Fig. 7g). Apparently, the depletion of macrophages had restricted impact on 20F2 remedy (Fig. 7h), indicating that macrophages and ADCP are usually not the main anti-leukemia effectors mediated by 20F2 in vivo. Since 20F2 may selectively block the HHLA2-independent TMIGD2 signaling in AML cells however largely keep costimulatory signaling mediated by HHLA2-TMIGD2 interplay on T and NK cells (Supplementary Fig. 7h, i), which is probably higher than 17C7, we handled extra AML PDX fashions that have been established with hard-to-treat or secondary AML affected person samples (Pt#19, Pt#8 and Pt#24), and located that every one responded effectively to 20F2 remedy (Fig. 7i). Subsequent, we examined 20F2 within the first spherical PDX fashions established with AML affected person samples representing three subsets of AML based mostly on TMIGD2 expression, together with CD34+TMIGD2low (Pt#31, <10% TMIGD2+ cells), CD34+TMIGD2excessive (Pt#28, >80% TMIGD2+ cells) and CD34–TMIGD2+ (Pt#21, 37% TMIGD2+ cells). The primary spherical PDX mice have been handled with isotype management versus 20F2. Downregulation of TMIGD2 and decrease proportion of CD34+CD38– cells have been detected in 20F2 handled group in contrast with isotype management (Supplementary Fig. 7j,ok). The second spherical NSG-recipient have been xenotransplanted intravenously with FACS-purified CD33+ myeloid cells remoted from the primary spherical AML PDX mice. The second spherical NSG-recipient acquired no additional 20F2 remedy. We noticed vital lowered leukemia burden in each rounds of PDX mice (Fig. 7j), indicating that anti-TMIGD2 mAb preferentially inhibit leukemogenesis by focusing on TMIGD2-expressing LSCs.
To check whether or not the sensitivity of regular HSPCs to 20F2 was the identical as AML samples, 20F2 was used to deal with NSG-recipients engrafted with CD34+ cells from twine blood. In distinction to the remark in AML PDX fashions, 20F2 confirmed negligible inhibitory results on the engraftment of regular cells, together with whole hCD45+ cells, hCD19+ lymphoid cells, and CD33+ myeloid cells (Fig. 7k, Supplementary Fig. 7l), which might be a consequence of comparatively low floor TMIGD2 expression and intrinsic relative TMIGD2-signaling independence in HSPCs. Taken collectively, focusing on TMIGD2 with mAb differentially eliminates AML cells over regular HSPCs, and is a promising therapeutic technique.