Cohesin-mutant hematologic malignancies have distinct illness traits
We investigated 2 giant cohorts of sufferers recognized with a hematologic malignancy for presence of a mutation within the cohesin advanced (Supplementary Fig. 1). In complete, we recognized 790 sufferers with a pathogenic mutation in any of the cohesin advanced genes (Fig. 1A). The incidence of cohesin mutation was 10% in MDS, 5% in MDS/MPN, and eight% in AML sufferers within the DFCI cohort (Supplementary Desk 2). Mutations in numerous cohesin subunits have been famous to be mutually unique with one another, with solely 12/790 (2%) instances characterised by mutations in additional than 1 cohesin subunit, and so they have been unfold all through the coding sequence with none hotspots (Fig. 1B and Supplementary Fig. 2). STAG2 mutations have been the most typical and current in 610 (77%) of sufferers, adopted by RAD21 in 104 instances (13%), SMC3 in 26 instances (3%), SMC1A in 22 instances (3%), and PDS5B in 16 instances (2%). Frameshift indel mutations have been probably the most frequent sort of mutations for many cohesin genes apart from STAG2, the place nonsense mutations resulting in a untimely cease codon comprised greater than 50% of all mutations (Supplementary Fig. 3A). Sufferers with mutations within the cohesin advanced have been recognized with AML, MDS, and MDS/MPN in 374, 351, and 63 instances, respectively (Desk 1, Supplementary Fig. 3B). Within the DFCI cohort, we recognized 55 sufferers (17.7%) with a pathologist-validated non-myeloid hematologic malignancy, together with indolent and high-grade lymphomas (persistent lymphocytic leukemia (n = 7), diffuse giant B cell lymphoma (n = 7), a number of myeloma (n = 3), and acute lymphoblastic leukemia (n = 8)) (Supplementary Fig. 3C). In all subsequent analyses, we excluded instances with non-myeloid hematologic malignancies and targeted on sufferers with AML, MDS, and MDS/MPN solely.
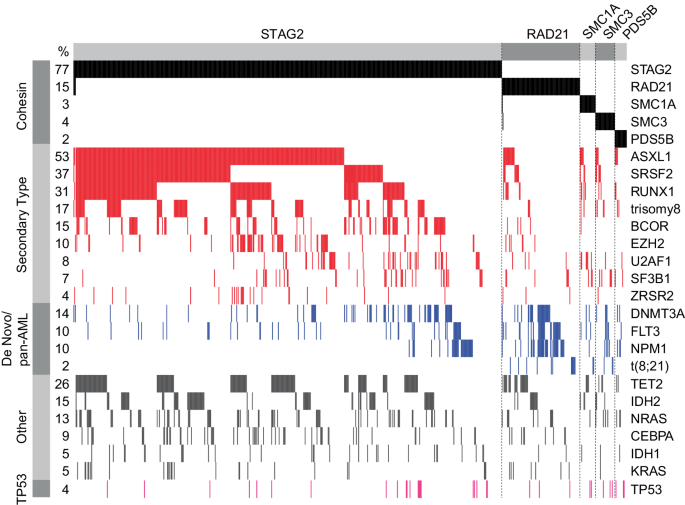
A Oncoprint of all sufferers with cohesin mutations (DFCI and MLL cohorts mixed), n = 790. B Lollipop plot panel of cohesin mutations for the mixed cohort. C Pie charts of distribution of cohesin mutations throughout MDS, AML, and MDS/MPN for the mixed cohort. D Field plot of the full variety of pathogenic mutations recognized by focused sequencing of sufferers with cohesin-mutant MDS and AML on the time of prognosis, stratified by cohesin standing for the mixed cohort. Wilcoxon take a look at was used to find out significance.
Sufferers have been divided right into a cohesin-WT (n = 4487) or cohesin-MT (n = 735) cohort. We then systematically in contrast the medical and demographic options of those 2 cohorts. Cohesin-MT sufferers have been older on the time of prognosis (AML: 69 vs. 65 years; MDS: 73 vs. 69 years; p < 0.0001) and had completely different patterns of AML and MDS subtypes than their cohesin-WT counterparts. AML with myelodysplasia-related defining genetic abnormalities (AML-MR) was current in 73% of cohesin-MT in comparison with 34% of cohesin-WT instances (p < 0.001). Conversely, AML with out genetically outlined lesions (summarized as AML by differentiation) and AML with NPM1 mutation have been extra frequent amongst cohesin-WT than cohesin-MT instances (31% vs. 8.3%, p < 0.001; 14 vs. 11%, p = 0.14, respectively). Inside MDS, the cohesin-MT cohort had a better fraction of extra superior MDS diagnoses than the cohesin-WT cohort (MDS-IB1: 35% vs. 16%, p < 0.0001; MDS-IB2: 36% vs. 17%, p < 0.0001). In keeping with these findings, the fraction of sufferers with documented development from MDS to AML was increased in MDS sufferers with cohesin mutations than MDS sufferers with out these mutations (32% vs. 21%, p = 0.005; knowledge solely obtainable for the DFCI cohort). Notably, MDS with bi-allelic TP53 inactivation, del5q, and SF3B1 related MDS have been almost mutually unique with cohesin-mutant MDS (Desk 1, Supplementary Desk 2). These knowledge display that cohesin mutations segregate with distinct medical options linked to MDS and subsequent secondary AML.
Cohesin advanced mutations have distinct medical options and AML ontogeny
We subsequent aimed to delineate variations amongst mutations of the cohesin advanced parts in sufferers with AML, MDS, and MDS/MPN overlap syndromes (Desk 2, Supplementary Desk 3). Given their excessive prevalence, our evaluation targeted on the comparability between instances with STAG2 versus RAD21 mutations, and examined the impression of SMC1A, SMC3, and PDS5B mutations as a gaggle on account of their considerably decrease numbers (thereafter known as SMC1A/SMC3/PDS5B). We noticed that sufferers with STAG2 mutations have been older than sufferers with mutations in different cohesin genes at time of AML prognosis (70 vs. 64 vs. 64/57/50 years, p < 0.001, Supplementary Fig. 3D) however not at time of MDS prognosis (Supplementary Fig. 3E). We noticed a major distinction within the distribution of cohesin subunit mutations, with 72.5% of all AML instances, however 87.4% of all MDS instances (p < 0.001) and 93% of MDS/MPN instances (p = 0.001) carrying a STAG2 mutation (Fig. 1C, Desk 2). Moreover, there was a major distinction within the AML ontogeny amongst completely different cohesin mutations. STAG2 mutations have been virtually solely related to an AML-MR prognosis (256/271, 94%), with uncommon instances of NPM1 and bi-allelic CEBPA (8/271 (3%) and 5/271 (1.8%), respectively) (Desk 2). Conversely, sufferers with RAD21 and SMC1A/SMC3/PDS5B mutations introduced extra steadily with de novo AML with NPM1 mutations (27/69 (39%), 2/12 (17%), 3/11(27%), and a couple of/6 (33%) instances, respectively) versus STAG2-mutant sufferers (p < 0.0001). Core binding issue leukemia t(8;21)/RUNX1::RUNX1T1 was present in 10/69 (14%) of RAD21-mutant sufferers and a couple of/29 (7%) of SMC1A/SMC3/PDS5B-mutant sufferers however solely in 1/271 (0.4%) STAG2-mutant affected person (p < 0.0001 and 0.026, Desk 2).
Constantly, RAD21 and SMC1A/SMC3/PDS5B mutations represented a considerably higher proportion of cohesin-mutant AML than MDS as in comparison with STAG2 mutations (Fig. 1C). This implies that STAG2 mutations are typically acquired on the MDS stage, and RAD21 and the opposite cohesin subunit mutations could also be extra possible acquired on the AML stage and result in speedy leukemic transformation relatively than a slower enhance in blast rely over time, as could also be anticipated in MDS. Certainly, sufferers with RAD21 and SMC1A/SMC3/PDS5B mutations trended in the direction of a better proportion of blasts of their diagnostic AML bone marrow biopsy in comparison with sufferers with STAG2 mutations (median morphology-defined blast rely of 47% for RAD21 vs. 28% for STAG2-mutant AML, p = 0.11, Supplementary Fig. 3F, knowledge obtainable for the DFCI cohort solely).
To additional examine whether or not STAG2 mutations could also be preferentially acquired at MDS stage and result in improvement of secondary AML, we extracted all sufferers with obtainable longitudinal mutation knowledge and recognized 23 sufferers recognized with STAG2-mutant AML with a minimum of one mutational evaluation earlier than AML prognosis. The median time from the primary mutational evaluation to AML prognosis was 13 months (vary 6–53 months) (Supplementary Fig. 4A, B and 5A). Out of 23 instances, 17 sufferers (73%) had a STAG2 mutation that was detected previous to AML prognosis, of which 13 sufferers (76%) have been recognized with MDS, and 4 sufferers (24%) with MDS/MPN. There have been 5/23 sufferers (22%) that confirmed the primary emergence of a STAG2 mutation at AML prognosis, and only one/23 sufferers (4%) acquired the STAG2 mutation upon AML illness relapse. Moreover, we noticed a comparatively steady STAG2 VAF main as much as AML prognosis (Supplementary Fig. 4C, p = 0.85), suggesting that the STAG2-mutant clone had already dominated the bone marrow in non-leukemic cells earlier than the AML prognosis was made. These knowledge collectively counsel that the medical presentation of sufferers with mutations in numerous cohesin subunits will not be uniform, with completely different mutations affecting illness biology and AML ontogeny in distinct methods. Moreover, our knowledge display that STAG2 mutations don’t act as AML-defining lesions and are normally acquired on the MDS stage.
Cohesin advanced mutations are related to distinctive co-mutational, cytogenetic and transcriptional profiles
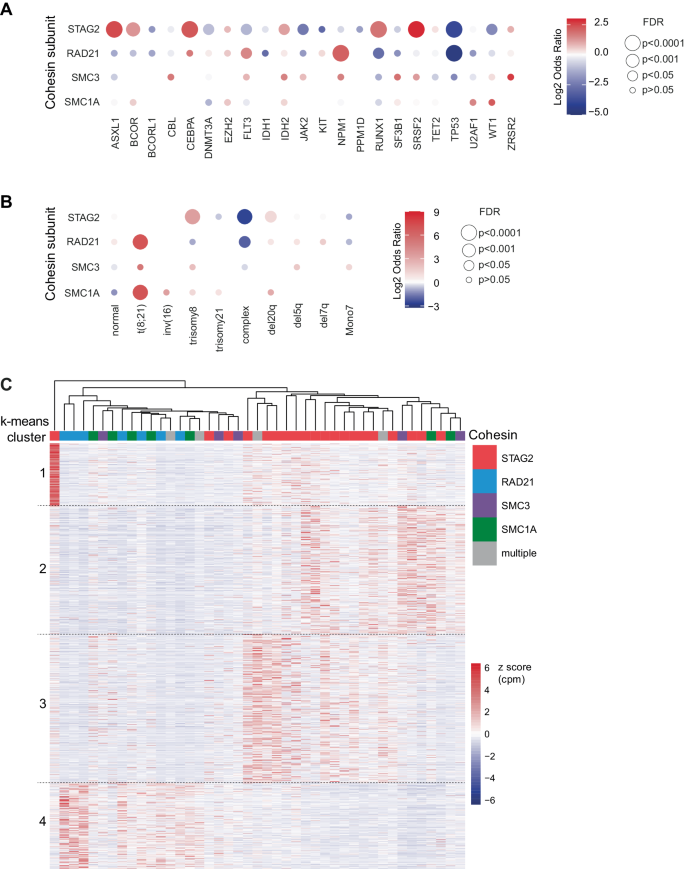
Having established a novel sample of illness traits of cohesin mutations, we subsequent examined the genetic traits related to every mutation. To dissect the genetic make-up of STAG2 and different cohesin gene mutations, we first in contrast the full variety of extra detected mutations. We noticed that sufferers with STAG2 mutations had a better variety of co-mutations in comparison with cohesin-WT sufferers (median 4 vs. 3, p < 0.001), and all different cohesin mutations (median 4 vs. 3, p < 0.003, Fig. 1D, Supplementary Fig. 3H). Subsequent, we analyzed variations within the co-mutational patterns between STAG2 and RAD21-mutant AML and MDS sufferers (Fig. 2, Supplementary Fig. 5–6). Essentially the most steadily co-occurring mutations with STAG2 have been secondary sort mutations, together with mutations in ASXL1 (64%), SRSF2 (45%) and RUNX1 (37%). In distinction, RAD21 mutations have been more likely to co-occur with de novo or pan-AML mutations, together with NPM1 (32%) and FLT3 (23%). We additionally noticed t(8;21) in 14% of RAD21-mutated instances, with and with out extra KIT mutations, however not in a single case of STAG2-mutant illness. By systematically evaluating the co-mutational panorama of STAG2 mutations and different cohesin gene mutations (Fig. 3A), we discovered a major optimistic enrichment of STAG2 being co-mutated with secondary ontogeny-defining mutations, together with ASXL1, SRSF2, BCOR, and RUNX1 (OR = 5.6, 7.5, 2.5, 3.5; false discovery charge (FDR)= 4×10–29, 5×10–31, 7×10–5, 2×10–13, respectively; Supplementary Desk 4, Supplementary Fig. 7A-B). Conversely, mutations in NPM1 and DNMT3A have been underrepresented in sufferers carrying a STAG2 mutation (OR = 0.7 and 0.5; FDR = 0.5 and 0.004, respectively). In distinction, the co-mutational sample for RAD21, SMC3, and SMC1A was distinct from STAG2, and confirmed enrichment for de novo and pan AML ontogeny-defining mutations NPM1 and FLT3 (OR = 4.4 and a couple of.3; FDR = 1×10–5 and 0.01, respectively). Curiously, STAG2 and RAD21 mutations shared a near-mutational exclusivity with TP53 mutations (OR = 0.04–0.06, FDR < 0.001), which was evident for MDS and AML instances (ORSTAG2-MDS = 0.07, ORSTAG2-AML = 0.05, ORRAD21-MDS = 0, ORRAD21-AML = 0.08).
Oncoprint for MDS, AML, and MDS/MPN sufferers from the mixed cohort. Circumstances with STAG2, RAD21, SMC3, SMC1A, and PDS5B mutations have been sorted by co-mutational sample based mostly on illness ontogeny [21] and affiliation with cohesin subunits. A 2% allelic frequency lower off was used.
Balloon plot for relative enrichment of co-occurrence of cohesin subunit mutations with different myeloid driver mutations (A) and chromosomal aberrations (B). Cohesin-WT cohort was used as reference to calculate enrichment, which is indicated as log2 odds ratio (OR). Mixtures with q < 0.05 or 5% mutational frequency within the complete cohort are proven in (A). Lacking balloons point out OR = 0. False discovery charge (FDR) is as indicated: ***<0.0001, **<0.001, *<0.05 and corresponds to dot measurement. C Gene expression heatmap of cohesin mutant sufferers with annotated cohesin mutations and clustered by k-means (1328 genes). Samples have been clustered by hierarchical clustering and annotated by cohesin mutation. Colour point out z rating reworked CPM per gene.
We equally noticed a definite sample of co-occurring cytogenetic aberrations amongst cohesin-MT instances (Fig. 3B, Supplementary Fig. 7C, D). Trisomy 8 was enriched in STAG2-mutant versus cohesin-WT instances (OR = 10.6, FDR = 2.2×10–50). In distinction, mutations in RAD21, SMC1A, and SMC3 have been enriched for t(8;21) (OR = 134, 131, 37; FDR = 4.1×10–19, 3.8×10–5, 0.14, respectively). Advanced karyotypes have been much less frequent amongst all cohesin-MT sufferers in comparison with cohesin-WT sufferers (ORSTAG2 = 0.12, FDR = 2.1×10–22; ORRAD21 = 0.24, FDR = 0.006), which might be anticipated given their near-mutual exclusivity with TP53 mutations.
Our genetic and cytogenetic analyses supported the speculation that STAG2- and non-STAG2- mutant myeloid illnesses symbolize distinct biology and ontogeny. To analyze whether or not this was supported by distinct gene expression packages, we analyzed transcriptomic profiles of the cohesin-mutant AML instances within the BeatAML cohort [36]. Utilizing unsupervised k-means clustering, we noticed distinct transcriptional profiles of STAG2– and RAD21/SMC3/SMC1A-mutant instances (Fig. 3C). Gene set enrichment evaluation (GSEA) highlighted differential expression of viral response and interferon signaling, in addition to metabolic packages and extracellular matrix-associated pathways (Supplementary Fig. 8). In abstract, the distinct co-mutational, cytogenetic and molecular landscapes of various cohesin mutations counsel distinctive patterns of illness improvement pushed by completely different cohesin mutations.
STAG2 mutations have prognostic impression in MDS and AML
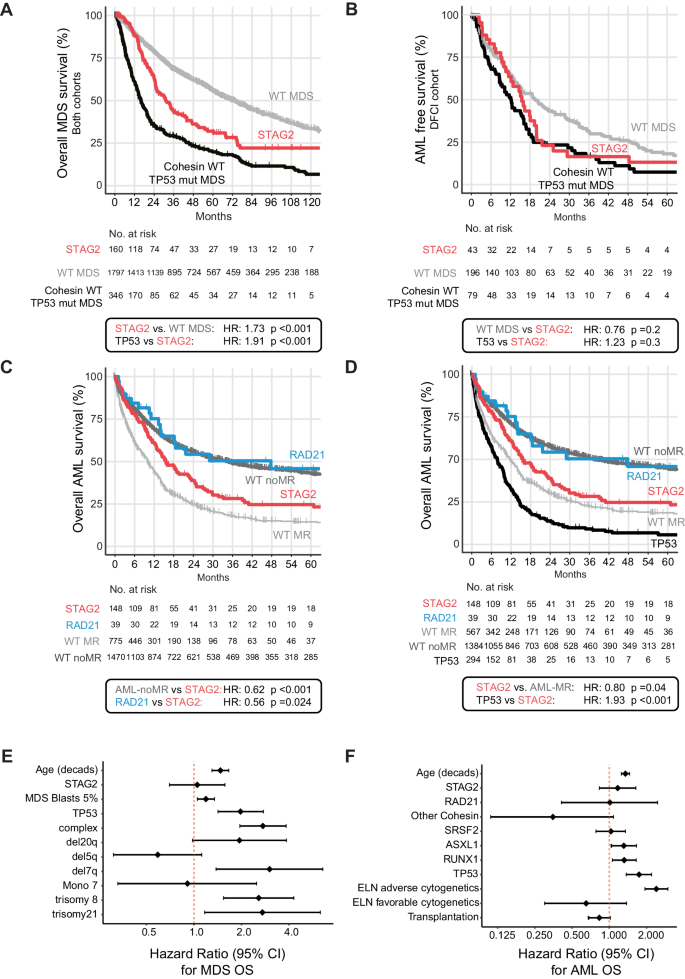
Having established distinctive illness and genetic traits for various cohesin mutations, we subsequent assessed their impression on medical outcomes. We performed unbiased analyses of general survival (OS) and development free survival (PFS) in MDS and AML. The median follow-up time for the complete affected person cohort was 73.8 months (95% confidence interval (CI) = 69.6–80.5 months) for MDS, and 49.4 months (95% CI = 45.4–54.4 months) for AML. We first in contrast outcomes for STAG2-mutant MDS to cohesin-WT MDS during which STAG2 conferred a poor danger at a median OS of 30.3 versus 58.9 months (HR: 1.44, 95% CI 1.17–1.78, p < 0.001, mixed cohort, Supplementary Fig. 9). Given our observations of near-mutual exclusivity of cohesin and TP53 mutations (Figs. 2 and 3A), and the well-established affiliation of TP53 mutations with poor outcomes [38, 39], we subsequent in contrast the OS and AML-free survival of sufferers with STAG2-mutant MDS to TP53-mutant MDS and cohesin/TP53-WT MDS. We noticed a considerably worse OS of STAG2-mutant MDS in comparison with cohesin/TP53-WT MDS (HR = 1.73, 95% CI = 1.4–2.14, median OS 30.3 vs. 69.8 months, p < 0.001, Fig. 4A), and an identical danger of leukemic transformation in STAG2- and TP53-mutant MDS instances (median AML-PFS of 15.4 months for STAG2 and 12.1 months for TP53, p = 0.3, DFCI cohort solely, Fig. 4B). In a multivariable regression evaluation to establish the impact of mutations, cytogenetics, diagnostic blast rely, and age on the time of MDS prognosis, the presence of a STAG2 mutation didn’t attain significance as an unbiased predictor of MDS consequence (Fig. 4E).
Survival evaluation utilizing the Kaplan-Meier methodology and log-rank take a look at for general survival in MDS (A), AML-Development free survival in MDS (DFCI cohort solely) (B), and AML survival stratified by cohesin subunit mutational standing and cohesin-WT group by AML MR or AML-non-MR (C) and TP53 mutation (D). HR = Hazard ratio. Statistical significance was decided utilizing the log rank-test. E Forrest plot for multivariate prognostic impression of STAG2 mutations for MDS OS and (F) for AML OS.
For our consequence evaluation in AML, we first in contrast STAG2-mutant AML to cohesin-WT instances separated into cohesin-WT AML related to myelodysplasia-related modifications (thereafter known as “AML-MR”) and cohesin-WT AML not related to myelodysplasia-related modifications (thereafter known as “AML-non-MR”) based on the WHO 2022 classification. We noticed that STAG2-mutant AML had considerably worse OS than AML-non-MR (HR = 0.62, 95% CI = 0.5–0.76, median OS 16 vs. 35 months, p < 0.001), and solely a modestly higher OS than AML-MR (HR = 1.43, 95% CI = 1.16–1.77, median OS 10.3, p < 0.001, Fig. 4C). Given the near-mutual exclusivity of STAG2 and TP53 mutations in AML, we carried out a subset evaluation of cohesin-WT AML excluding TP53-mutant instances which eliminated many of the variations and confirmed a really comparable and numerically even favorable consequence between STAG2-mutant and AML-MR with out TP53 mutations. (HR = 0.80, 95% CI = 0.64–0.99, median OS 13.6 vs. 11.8 months, p = 0.04, Fig. 4D). This poor consequence was additionally evident for the uncommon STAG2-mutant instances that weren’t recognized as AML-MR due to competing classifying mutations (e.g., NPM1 and/or CEPBA, Supplementary Fig. 10A, B).
Importantly, this sample was distinct from the outcomes of RAD21-mutant AML, which was virtually equivalent to AML-non-MR and considerably higher than STAG2-mutant AML OS (HR = 0.56, 95% CI = 0.34–0.93, median OS 48 vs. 16 months p = 0.024) (Fig. 4C, Supplementary Fig. 11A, B). This impact was most obvious within the DFCI cohort, though we noticed the identical development within the MLL cohort, with variations possible being pushed by intrinsic variability in remedy and choice biases between DFCI and MLL (Supplementary Fig. 11C, D). Allogenic stem cell transplantation instances accounted for 41% of DFCI however solely 12% of MLL instances (Supplementary Desk 5), and response charges to induction remedy (Supplementary Desk 6) have been comparable between teams. The results of STAG2 and RAD21 mutations on OS remained important when censored for allogeneic stem cell transplantation (Supplementary Fig. 10B, Supplementary Fig. 12), though neither one reached statistical significance as an unbiased predictor of consequence in a multivariable regression evaluation of identified medical co-variables (age and transplantation as time dependent variables) and co-mutation with ASXL1, SRSF2, RUNX1 and TP53 (Fig. 4F).
In abstract, our findings counsel that solely STAG2 mutations confer a detrimental impression on AML outcomes, which is attributed to secondary ontogeny and a genetic make-up of previous myeloid dysplasia. Notably, the prognostic impression of RAD21 mutations is shared with de novo AML.