Identification of main cell sorts
To systematically consider TME significance in CRC sufferers, we obtained scRNA-seq knowledge from 3 datasets, encompassing 35 CRC sufferers and 4 wholesome controls, with 34 tumor and 23 non-tumor samples. Among the many 23 non-tumor samples, 19 had been adjoining non-malignant tissues from CRC sufferers. To attenuate batch impact among the many scRNA-seq datasets, we individually assessed particular person dataset. Following low-quality cell filtration, we acquired 42,696, 68,702, and 118,904 single cells, and generated unsupervised clustering of 24, 31, and 39 clusters, respectively for the GSE200997, GSE166555 and GSE201348 CRC datasets (Fig. 1A, Supplementary Figs. 1A and 2A). The aforementioned clusters had been, in flip, separated into 7 major cell elements, in response to their related canonical markers, and these had been, fibroblasts (harboring ACTA2, MCAM, MYLK, MYL9, FAP and THY1), epithelial cells (harboring EPCAM, SFN, KRT19, KRT18 and CDH1), myeloid cells (harboring CD68, CD14, FCGR3A,LYZ and MARCO), endothelial cells (harboring VWF, PECAM1 and CD34), NK/T cells (harboring NKG7, GNLY, KLRD1, CD24, CD3,CD4 and CD8), B cells (harboring CD19, MS4A1,PAX5 and CD79A) and plasmaB cells (harboring MZB1, JCHAIN, JCHAIN and SDC1) (Fig. 1B,D, Supplementary Figs. 1B,D and 2B,D). Determine 1C, Supplementary Figs. 1C and 2C illustrate the cell-type compositions and tissue origins of every dataset. Primarily based on our commentary, cells from tumor samples had been built-in with cells from non-tumor samples. This indicated no marked batch results amongst totally different samples throughout clustering. To raised elucidate mobile clustering, we carried out cell proportion evaluation. We revealed un-even distribution of the relative abundances of the 7 major mobile populations between tumor and non-tumor cells (Fig. 1E, Supplementary Figs. 1E, and 2E). Given this proof, it was clear that the TME strongly modulated standing of CRC sufferers.
A abstract of the only cells in CRC sufferers, and recognition of major cell sorts within the GSE200997 dataset. (A) UMAP plot depicting single cells (coloured in response to cell cluster). (B) UMAP plot depicting single cells (coloured in response to mobile kind). (C) UMAP plot depicting single cells (coloured in response to pattern origins, both tumor versus regular samples). (D) Dot plot illustrating consultant marker genes throughout all mobile clusters. Dot dimension signifies fraction of particular gene-expressing cells. Coloration depth signifies relative particular gene expressions. (E) Stacked bar chart depicting 7 main mobile kind contents in particular person tumor or regular samples.
From the three aforementioned datasets, we respectively acquired 25,667, 38,233, and 17,713 cells from tumor tissues. Earlier research have revealed T13, myeloid7 and fibroblast15 cells performed an vital position within the development of CRC. Thereafter, to higher elucidate the T, myeloid and fibroblast profiles in numerous TME of CRC LI sufferers in larger element, we mixed these cells for tumor tissues from all 3 datasets, particularly, NK/T, myeloid, and fibroblasts cell populations. Following batch impact correction, we carried out second stage clustering, and we obtained range of cell populations (Figs. 2C, 3A, and 4A). This confirmed that there have been complicated TMEs in CRC sufferers. Lastly, to discover the TMEs-mediated regulation of LI in CRC sufferers, we examined the hyperlink between cells and LI-specific phenotypes. Our findings from the aforementioned analyses are described intimately under (Figs. 2D, 3B, and 4B).
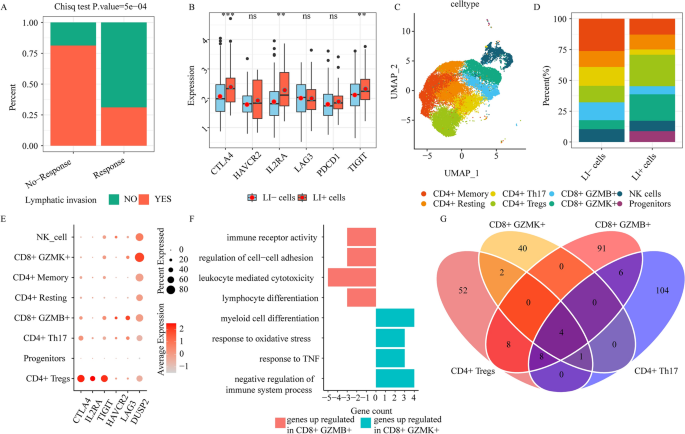
Immune response is diminished in lymphatic invasion (LI) sufferers. (A) Bar graph depicting the first therapeutic response (full/partial response (response), steady/progressive illness (no-response)) for LI and no-LI TCGA sufferers. (B) The immune checkpoint gene expression profiles between LI- and LI+ cells (ns ≥ 0.05, * < 0.05, ** < 0.01, *** < 0.001 and **** < 0.0001). (C) UMAP plots depicting all NK and T cells, coloured in response to cell sub-clusters. (D) Stacked bar chart depicting detailed elements of particular person NK/T cell clusters in LI- or LI+ cells. (E) The immune checkpoint gene expressions in 8 NK/T cell subclusters. (F) Enrichment evaluation of upregulated genes in CD8+ GZMK+ or CD8+ GZMB+ cells. (G) The venn plot depicting DEG contents in CD4+ Tregs, CD8+ GZMK+, CD4+ Th, and CD8+ GZMB+, by evaluating LI+ or LI-cells to the remaining cells.
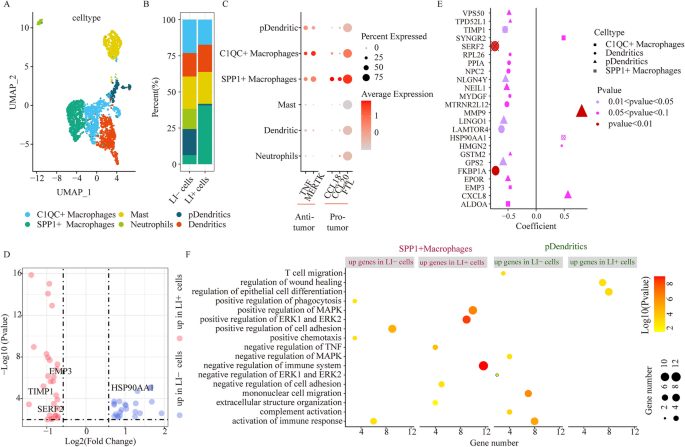
Myeloid mobile cluster comparability between LI+ and LI- cells. (A) UMAP plots depicting all myeloid cells, coloured in response to mobile sub-clusters. (B) Stacked bar chart illustrating the detailed compositions of particular person myeloid cell clusters in LI- and LI+ cells. (C) The tumor suppressor and tumor selling gene expressions in particular person myeloid sub-clusters. (D) DEG analysis by way of comparability of LI+ or LI-cells to the remaining cells in SPP1+ macrophages. (E) Affiliation between gene expression and OS for DEGs by way of comparability amongst LI+ or LI-cells and the remaining cells in particular person myeloid sub-clusters. Dot dimension represents absolutely the correlation coefficient worth, and form signifies the cell subsets. (F) Enrichment evaluation for DEGs by way of comparability of LI+ or LI-cells to the remaining cells in SPP1+ macrophages and pDCs.
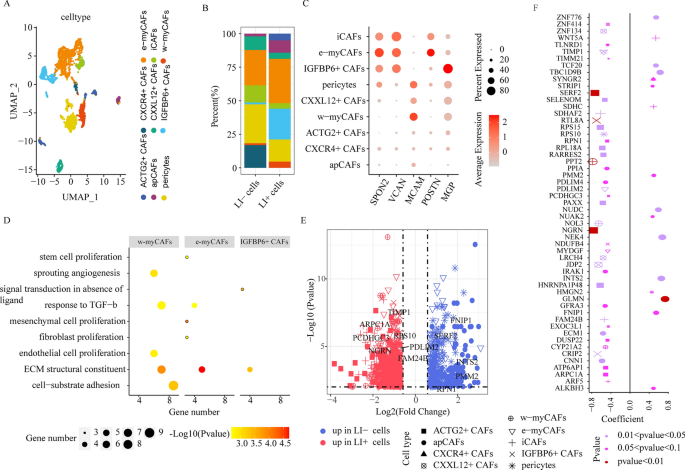
CAF cell cluster comparisons between LI+ and LI- cells. (A) UMAP plots depicting all CAF cells, coloured in response to the cell sub-clusters. (B) Stacked bar chart illustrating the detailed compositions of particular person CAF cell clusters in LI- and LI+ cells. (C) The expression profiles of genes (SPON2, VCAN, MCAM, MGP, and POSTN) whose enhanced expression is correlated with illness development and metastasis, in particular person CAF sub-clusters. (D) Enrichment analyses of considerably upregulated genes in e-myCAFs, w-myCAFs, and IGFBP6+ CAFs. (E) DEG analysis by way of comparability of the LI+ or LI-cells to the remaining cells in particular person CAF cell clusters. (F) Affiliation between gene expression and affected person OS for DEGs by way of comparability of LI+ or LI-cells to the remaining cells in particular person CAF sub-clusters. Dot dimension represents absolutely the correlation coefficient worth, and form signifies the mobile subsets.
Distinct CD8+ and CD4+ T cell states regulate the pro-invasive immune response
We retrieved the TCGA-COAD clinicopathological knowledge (comparable to, lymphatic_invasion and first remedy final result success) from UCSC xena, which examined 164 LI sufferers and 250 no-LI sufferers, together with 195 full response sufferers, 12 partial response sufferers, 25 progressive illness sufferers, and 4 steady illness sufferers. Our in depth evaluation of LI and first therapeutic success revealed that LI sufferers exhibited considerably diminished therapeutic response and skilled worse survival, relative to no-LI (Fig. 2A, Supplementary Fig. 3). To raised elucidate the single-cell transcriptome knowledge between LI and no-LI sufferers, we employed scissor to correspond single-cell knowledge to LI-specific phenotypes by way of TCGA-COAD bulk RNA-seq pattern. We revealed 513 LI-related (LI+ cells) and 641 no-LI-related (LI− cells) cells (Supplementary Fig. 4A). Notably, immune checkpoint genes, comparable to, CTLA4, IL2RA, and TIGIT had been strongly differentially expressed (DE) between LI+ and LI− cells (Fig. 2B). To discover the position of T cells in selling LI formation in CRC sufferers, we stratified NK/T cells into progenitors, NK cell, CD4+ T cell, and CD8+ T cell clusters (Fig. 2C, Supplementary Fig. 4B). Particular person cell clusters exhibited contributions from distinct datasets (Supplementary Fig. 4C), which indicated no marked batch results throughout clustering. Among the many CD4+ T cell states, CD4+ Tregs (harboring FOXP3), CD4+ Reminiscence (harboring CCR7, SELL, and TCF7), CD4+ Resting (harboring ANXA1), and CD4+ Th (harboring IL22 and IL17A) had been acknowledged as 4 impartial clusters (Supplementary Fig. 4B). Of be aware, we demonstrated that CD4+ Tregs had been extra enriched in LI+ than in LI− cells (Fig. 2D). We additional validated this utilizing CD4+ Tregs signature gene set scores by way of the TCGA-COAD bulk RNA-seq (Supplementary Fig. 4D). Extra evaluation revealed that the immune checkpoint markers, CTLA4, IL2RA, and TIGIT, had been comparatively considerable in CD4+ Tregs (Fig. 2E). Primarily based on these evidences, the tumor immune microenvironment (TIME) strongly contributes to shaping LI, which is, in flip, regulated by immune evasion throughout LI states in CRC sufferers.
Among the many CD8+ T cell states, CD8+ GZMB+ and CD8+ GZMK+ had been acknowledged as 2 impartial CD8+ cytotoxic cell clusters (Fig. 2C, Supplementary Fig. 4B). These mobile states had been beforehand reported in CRC sufferers13. CD8+ GZMK+, which possesses pro-inflammatory properties16, exhibited the next inhabitants in LI+ cells, relative to the LI- cells (Fig. 2C), and this was additional validated by CD8+ GZMK+ cell signature gene set scores utilizing the TCGA-COAD bulk RNA-seq (Supplementary Fig. 4D). Alternately, CD8+ GZMB+ cells had been enriched in LI− cells (Fig. 2C), which advised a doable antagonistic perform between CD8+ GZMK+ and CD8+ GZMB+ cells. Furthermore, we noticed no marked alterations within the CD8+ GZMB+ cell populations within the LI+ and LI− TCGA-COAD sufferers, which can be attributable to an underestimation of CD8+ GZMB+ cells infiltration abundance by bulk RNA-seq. To additional elucidate the antagonistic property between CD8+ GZMK+ and CD8+ GZMB+ cells, we carried out gene expression and Gene Ontology (GO) time period evaluation for the two CD8+ subsets. We revealed that the DUSP2 (serving as a T cell suppressor to attenuate host antitumor immunity17) expression in addition to the damaging immune system regulatory pathways had been considerably upregulated in CD8+ GZMK+ cells (Fig. 2E,F). In distinction, we revealed marked elevation in exhaustion marker expressions (LAG3, HAVCR2) in addition to the leukocyte-driven cytotoxicity pathway in CD8+ GZMB+ cells (Fig. 2E,F). Collectively, these evidences confirmed that TIME strongly regulated LI, and CD8+ GZMK+ cells accelerated LI by weakening the immune response.
We subsequent examined the transcriptome alterations in LI+ versus LI− states, and recognized differentially expressed genes (DEGs) in CD4+ Tregs, CD8+ GZMK+, CD4+ Th, and CD8+ GZMB+ cells. Primarily based on our commentary, most DEGs weren’t widespread amongst T cell clusters (Fig. 2G, Supplementary Fig. 4E,F). This advised that distinct molecular networks had been activated amongst distinct T cell sorts to advertise or inhibit LI.
Myeloid subtyping and contribution to LI
Prior investigations additionally reported that myeloid cells critically regulated CRC development7. Herein, we recognized 2603 myeloid cells, which had been sub-classified into 6 clusters, in response to their canonical marker gene expression, and these included macrophages (harboring MARCO, CD68, TREM2 and MRC1; together with 2 subsets), neutrophils (harboring CSF3R, S100A8 and S100A9), dendritic cells (harboring CD1C, CD1E, FCER1A and PKIB), pDCs (harboring DNASE1L3 and LAMP5) and mast cells (harboring MS4A2 and TPSAB1) (Fig. 3A,B and Supplementary Fig. 5A). We additionally recognized two macrophage subcategories, particularly, C1QC+ macrophages and SPP1+ macrophages utilizing upregulated expressions of C1QC and SPP1 respectively. Moreover, chemokine genes CCL1818 and CCL2019 are reported to speed up most cancers development by way of optimistic regulation of migration and angiogenesis, respectively, and these genes exhibited enhanced expression in SPP1+ macrophages (Fig. 3C). FTL20, which possesses each pro-proliferative and pro-angiogenic properties, was additionally upregulated in SPP1+ macrophages (Fig. 3C). In all our expression evaluation supported that the SPP1+ macrophages managed the pro-metastatic habits of CRC, which corroborated with prior studies7. Nonetheless, the C1QC+ macrophages which have been reported to carry out cytophagic and antigen-presenting perform7, exhibited enhanced TNF and MERTK expressions, which can decelerate tumor development (Fig. 3C). Collectively, our evidences relating to the 2 macrophage subsets carefully mirrored studies from earlier publication7 that existed the dichotomous purposeful phenotypes between SPP1+ macrophages and C1QC+ macrophages within the CRC TME7.
We subsequent examined the affiliation between relative abundance of assorted myeloid subtypes and LI-associated cells. Utilizing scissor, we acknowledged 293 cells associated to LI (LI+ cells), and 272 cells associated to no-LI (LI− cells) (Supplementary Fig. 5B). Extra importantly, we found that the SPP1+ macrophage inhabitants was augmented in LI+ versus LI− cells. In distinction, we revealed that plasmacytoid dendritic cells (pDCs) had been extra distinguished in LI− cells (Fig. 3B). Lastly, utilizing bulk dataset integration, we additionally achieved the identical conclusions (Supplementary Fig. 5C). We additional explored the differing transcriptional distributions of SPP1+ macrophages and pDCs between LI+ cells and LI− cells.
In case of SPP1+ macrophages, 28 genes had been extremely expressed in LI+ cells, whereas, 23 had been augmented in LI− cells (Fig. 3D). Of be aware, among the many elevated genes in LI+ cells, we noticed that the TIMP1 and SERF2 contents carefully related to poor survival of CRC sufferers (Fig. 3D,E). Moreover, we utilized the TCGA-COAD bulk samples to acquire the identical conclusion that TIMP1 and SERF2 contents had been elevated in LI sufferers (Supplementary Fig. 5D). Subsequently, we employed purposeful enrichment evaluation to point out that invasive networks, particularly, damaging immune modulation, damaging TNF regulation, optimistic MAPK modulation, optimistic ERK1, ERK2, and extracellular matrix (ECM) regulation, had been upregulated within the LI+ cells of SPP1+ macrophages (Fig. 3F). Alternately, among the many elevated genes in LI− cells, we noticed marked activation of immune response. In pDCs, 305 genes had been augmented in LI+ cells, and 46 genes had been augmented in LI− cells (Supplementary Fig. 5E). Among the many elevated genes in LI+ cells, we discovered a powerful inverse relationship between EPOR, GPS2, GSTM2, LINGO1, NEIL1, NLGN4Y, TPD52L1, and VPS50 gene expressions and affected person survival (Fig. 3E). Among the many elevated genes in LI-cells, the CXCL8 and MMP9 expressions had been positively related to affected person survival (Fig. 3E). Utilizing purposeful enrichment evaluation, we additional validated that the immune response-associated networks, particularly, IL-17, TLRs, and TNF axes had been strongly diminished in LI+ cells, in relation to LI− cells (Fig. 3F, Supplementary Fig. 5F). Collectively, these evidences indicated that the SPP1+ macrophage infiltration in LI+ cells accelerated LI, in flip pDCS infiltration in LI- cells, modulated anti-tumor immunity. Thus, concentrating on these signatures might diminish invasion and improve affected person scientific final result.
CAFs exert numerous capabilities in LI
Most cancers-associated fibroblasts (CAFs) are essential contributors to the TME. Following re-clustering, we recognized 9 fibroblast subsets with distinct properties, all of which had been outlined as CAFs owing to the expressions of angiogenesis- and immunomodulation-related genes (PDGFRA, PDGFRB, FAP, NOTCH3, HES4, THY1, CXCL12, CXCL14, CCL2, CXCR4, and ACTG2) (Supplementary Fig. 6). Subcluster cells that expressed antigen-presenting molecules, particularly, HLA-DRA and HLA-DRB1 had been designated as apCAFs (Fig. 4A, Supplementary Fig. 7A); cells that expressed collagen-associated genes, particularly, COL1A1, COL5A1, and ATCA2 had been known as myCAFs (Fig. 4A, Supplementary Fig. 7A); cells that expressed pericyte markers, particularly, RGS5, PDGFRB, and CD248, had been famous as pericytes (Fig. 4A, Supplementary Fig. 7A); and lastly, cells that expressed chemokines, particularly, CXCL12, CXCL14, and CCL2, had been acknowledged as iCAFs (Fig. 4A, Supplementary Fig. 7A). Among the many myCAF cell states, e-myCAFs (harboring ECM molecules like MMP14 and LOXL2) and w-myCAFs (harboring contractile myofibroblast elements, particularly, MYL9 and TAGLN) had been designated as two impartial clusters (Fig. 4A, Supplementary Fig. 7A). The remaining 4 subclusters had been outlined as CXCL12+ CAFs, CXCR4+ CAFs, ACTG2+ CAFs, and IGFBP6+ CAFs, respectively, primarily based on their distinctive gene expressions. This mirrored the CAF heterogeneity amongst CRC sufferers (Fig. 4A, Supplementary Fig. 7A).
Until date, there aren’t any single-cell degree research on CAF heterogeneity and affiliation with LI in CRC sufferers. Utilizing scissor, we recognized 218 cells as LI+ cells, attributable to their marked affiliation with LI, and 148 cells as LI− cells owing to their marked affiliation with no-LI (Supplementary Fig. 7B). Importantly, we revealed that each e-myCAFs and w-myCAFs had been extra considerable in LI+ versus LI− cells (Fig. 4B). Following a mixed evaluation of bulk datasets, we additionally obtained the identical conclusion that the e-myCAF and w-myCAF cell abundance was considerably upregulated in LI versus no-LI sufferers (Supplementary Fig. 7C). Furthermore, utilizing enrichment evaluation of considerably upregulated e-myCAF and w-myCAF genes, we revealed that each cells expressed elevated ranges of proliferation- and ECM transforming network-related genes, whereas, w-myCAFs additionally confirmed enrichment for angiogenesis-associated genes (Fig. 4D). Primarily based on these evidences, the upregulated e-myCAFs and w-myCAFs in CRC sufferers probably improve invasion and metastasis. Moreover, we revealed that invasion-related gene expression SPON221, VCAN22, and POSTN23 was closely upregulated in e-myCAFs, whereas, MCAM was augmented in w-myCAFs (Fig. 4C). Collectively, these outcomes advised that each e-myCAFs and w-myCAFs improve invasion and metastasis24. Furthermore, we revealed {that a} beforehand unreported CAF subpopulation (harboring IGFBP6, a gene strongly related to VSMC physiological perform25) (Fig. 4B), was extra enriched in LI+ cells than in LI− cells. Lastly, following a mixed evaluation of bulk datasets, we obtained the comparable conclusions (Supplementary Fig. 7C). Moreover, we revealed that the MGP expression, a gene that’s inversely associated to affected person prognosis, was considerably elevated in IGFBP6+ CAFs (Fig. 4C). To evaluate doable perform of augmented IGFBP6+ CAF abundance in CRC sufferers, we carried out pathway enrichment evaluation, and revealed that the ECM and sign transduction networks in absence of ligand had been remarkably enriched (Fig. 4D). Collectively, these evidences indicated that IGFBP6+ CAFs might speed up metastasis and modulate prognosis of CRC sufferers.
Moreover, the apCAF inhabitants, that are identified to control immune evasion in pancreatic most cancers26, was additionally remarkably enhanced in LI+ versus LI− cells, and this was additional validated utilizing apCAF signature gene set scores by way of TCGA-COAD-based bulk RNA-seq (Supplementary Fig. 7C). The apCAFs are additionally thought-about to be antigen presenting cells. Thus, we analyzed its affiliation with T cells. We revealed that, within the SPP1 axis-based interactions, apCAFs with enhanced SSP1 expressions exhibited extra mobile crosstalk with T cells with enhanced CD44 expression (Supplementary Fig. 7D). Because it has been early reported that the SPP1-CD44 ligand-receptor pair causes immunosuppression in intrahepatic cholangiocarcinoma development27, we speculated that the augmented apCAF abundance in CRC sufferers might probably induce immune escape. Collectively, these evidences advised that the heterogeneous CAFs exerted a number of capabilities to kind LI, together with accelerating invasion and enhancing immune escape.
Institution of three subcategories in CRC sufferers utilizing invasive genes
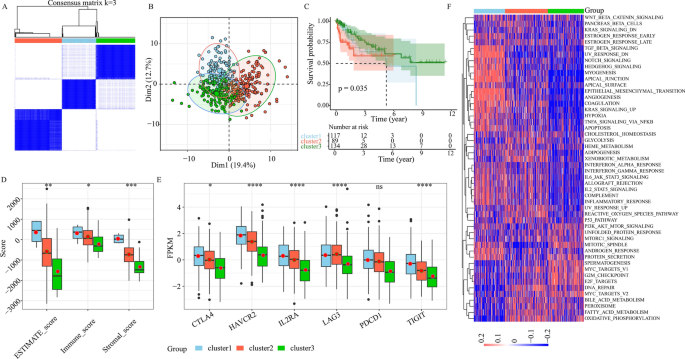
Herein, we employed single-cell and TCGA evaluation to judge the TME infiltration standing of CRC sufferers, and confirmed our identification of the LI-related cells from single-cell knowledge with phenotypic steering from bulk knowledge. This info can probably improve cell-targeted therapies and identification of sturdy prognostic markers. Via our comparability of LI+ or LI− cells with all different cells from particular person mobile subset, we carried out in depth DE (Fig. 4E) and general survival (OS) evaluation. Our conclusions had been as follows: 60 genes had been considerably elevated in LI+ cells, and had damaging correlation with affected person OS. Furthermore, 19 genes had been enhanced in LI− cells, confirmed optimistic correlation with affected person OS (Figs. 3E and 4F, and Supplementary Fig. 4F). Moreover, our unsupervised clustering evaluation of 79 gene expressions from the TCGA-COAD dataset revealed 3 distinct regulatory patterns. These included 129 circumstances in cluster 1, 178 circumstances in cluster 2, and 148 circumstances in cluster 3 (Fig. 5A). We subsequent separated all contributors into 3 cohorts, primarily based on the PCA outcomes, and confirmed the presence of three distinct subtypes (Fig. 5B). Primarily based on our survival evaluation, the three newly segregated subtypes exhibited markedly totally different prognosis (Fig. 5C, P < 0.05), with the cluster 3 sufferers experiencing the most effective prognosis.
Consensus clustering of lymphatic invasion (LI)-associated genes in TCGA-COAD. (A) Consensus matrices of TCGA sufferers. (B) PCA evaluation of the three subgroups in TCGA cohort. (C) KM curves depicting prognosis of the three TCGA subgroups. (D) Stromal, immune, and estimate scores among the many 3 subgroups (ns ≥ 0.05, * < 0.05, ** < 0.01, *** < 0.001 and **** < 0.0001). (E) The immune checkpoint gene expressions amongst 3 subgroups (ns ≥ 0.05, * < 0.05, ** < 0.01, *** < 0.001 and **** < 0.0001). (F) Heatmap depicting gene set variation evaluation scores of the 50 hallmark gene units within the 3 subgroups of colorectal most cancers (CRC). Coloration depth represents scores.
As well as, we carried out an intensive evaluation of clinicopathological profiles among the many 3 distinct subtypes. We noticed no discernible variations within the tumor stage, gender, age and stage among the many 3 subtypes (Supplementary Fig. 8). Nonetheless, the cluster 3 sufferers exhibited the very best proportion of no-LI (Supplementary Fig. 8). To raised elucidate variations in immune response, we in contrast the immune scores of assorted subtypes. We revealed that cluster 3 had the bottom immune cell and stromal cell scores, whereas, cluster 1 produced the biggest immune cell and stromal cell scores (Fig. 5D). We subsequent in contrast the profiles of immune checkpoint molecules, particularly, PD-1, CTLA4, LAG3, TIGIT, IL2RA, and HAVCR2. We revealed that the cluster 3 sufferers produced the bottom expressions of immune checkpoint molecules (Fig. 5E). Lastly, we demonstrated important variations within the carcinogenic profiles of the three clusters utilizing GSVA evaluation with the 50 Hallmark gene units. Cluster 1 confirmed enrichment within the EMT networks, particularly, TGF-B, NOTCH, and epithelial mesenchymal transition axes (Fig. 5F). Cluster 2 confirmed enrichment in metabolism networks, particularly, GLYCOLYSI and XENOBIOTIC (Fig. 5F). Cluster 3 confirmed enrichment in proliferation networks, particularly, E2F TARGETS, G2M CHECKPOINT, and MYC TARGETS (Fig. 5F). Primarily based on these evidences, we recognized sure LI targets, which modulate tumor growth, metastasis and immune response, and have nice potential in enhancing prognosis of CRC sufferers.
Improvement and validation of the LI-related prognostic mannequin
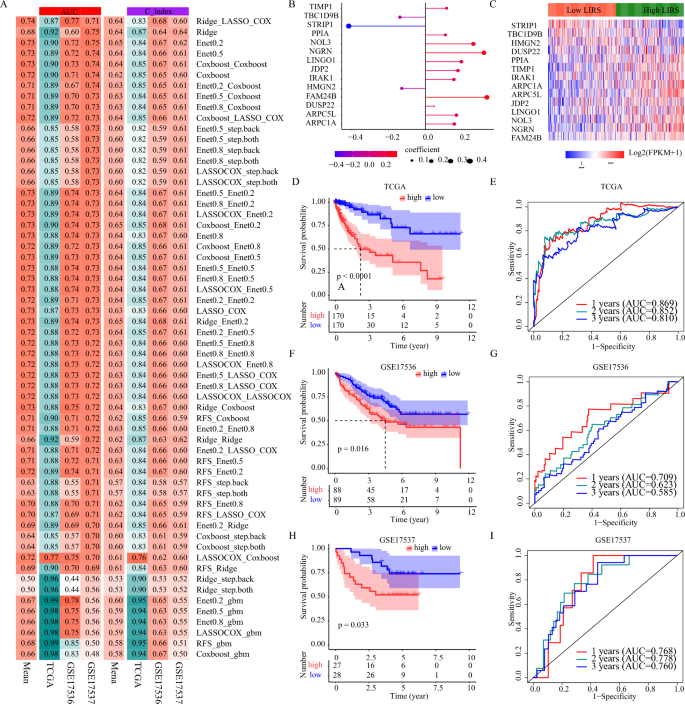
To develop a super biomarker for precisely stratifying the prognosis, primarily based on these above-mentioned LI targets, we utilized 60 machine-learning algorithm mixtures to assemble prediction fashions within the TCGA-COAD coaching cohort, and calculated the imply AUC and C-index of every algorithm within the two testing cohorts (GSE17536, GSE17537). As proven in Fig. 6A, the mix of Ridge (genes, with the coefficient > 0.01, had been chosen) and LASSO Cox with the very best common AUC (0.74) was chosen as the ultimate mannequin. As illustrated in Fig. 6B, the LI-related threat rating (LIRS) was developed in response to the expression of 14 LI-related signatures with following equation: LIRS = (0.168 ∗ ARPC5L) + (0.260 ∗ NOL3) + (0.115 ∗ TIMP1) + (0.334 ∗ FAM24B) + (0.108 ∗ PPIA) + (0.157 ∗ ARPC1A) + (0.317 ∗ NGRN) + (0.047 ∗ DUSP22) + (0.155 ∗ IRAK1) + (− 0.416 ∗ STRIP1) + ( − 0.138 ∗ TBC1D9B) + (0.177 ∗ JDP2) + ( − 0.128 ∗ HMGN2) + (0.195 ∗ LINGO1). The expression variations of the 14 LI-related signatures between excessive and low LIRS teams in response to the median worth in TCGA-COAD sufferers was proven in Fig. 6C. To guage the prognostic efficiency of LIRS, the Kaplan–Meier curve of OS demonstrated the excessive LIRS group possessed considerably shorter survival time within the TCGA-COAD coaching cohort (p < 0.0001 Fig. 6D). The time-dependent ROC curves at 1, 2 and three years of OS with the AUC values of 0.869, 0.852 and 0.810 confirmed the prognostic worth of the LIRS within the TCGA-COAD coaching cohort (Fig. 6E). The LIRS was additional independently validated within the two testing cohorts. The sufferers with excessive LIRS group possessed considerably shorter survival time each within the GSE17536 cohort and within the GSE17537 cohort (p < 0.05 Fig. 6F,H), and the AUC values of 1, 2 and three years had been 0.709, 0.623, and 0.585 within the GSE17536 cohort, 0.768, 0.778, and 0.760 within the GSE17537 cohort (Fig. 6G,I). The above outcomes additional clarified the LIRS might produce an correct prognostic prediction, and we offered priceless mechanistic insights into LI in scientific course of.
Improvement and validation of the LI-related prognostic mannequin. (A) The AUC and C-indexes of 60 machine-learning algorithm mixtures within the TCGA-COAD coaching cohort and the 2 testing cohorts. (B) Coefficients of the 14 LI-related signatures within the cox regression mannequin. (C) The differential expression of 14 LI-related signatures between high- and low-LIRS subgroups primarily based on median degree of LIRS in TCGA-COAD. (D–I) Kaplan–Meier survival curve of OS between high- and low-LIRS, and ROC curves at 1, 2 and three years within the TCGA-COAD coaching cohort and the 2 testing cohorts.