Creating mutation bushes
Three focused single-cell AML DNA sequencing datasets had been merged [10,11,12], leading to 207 sufferers with AML who had at the least two driver mutations, 275 samples, 823 mutation occasions, and 1 639 162 cells (Supplementary Fig. 1). Most circumstances had been cytogenetically regular (54%), and datasets had related affected person demographics however assorted in distributions of laboratory values, remedy, and pattern availability at prognosis (Desk 1). Though all sequencing panels lined 19 generally mutated genes, datasets differed within the dimension of the sequencing panels (Supplementary Desk 1), variety of cells per pattern, and the Tapestri Pipeline allele dropout estimate (Supplementary Fig. 6). After aggregating these datasets, mutations had been represented in related proportions as within the TCGA [21] and BeatAML [22] research, aside from enrichment in mutations widespread in AML, akin to in NPM1 and FLT3, and in low-level signaling mutations (Supplementary Fig. 7A); as an example, 58% of KRAS mutations had been in <10% of the corresponding pattern’s cells.
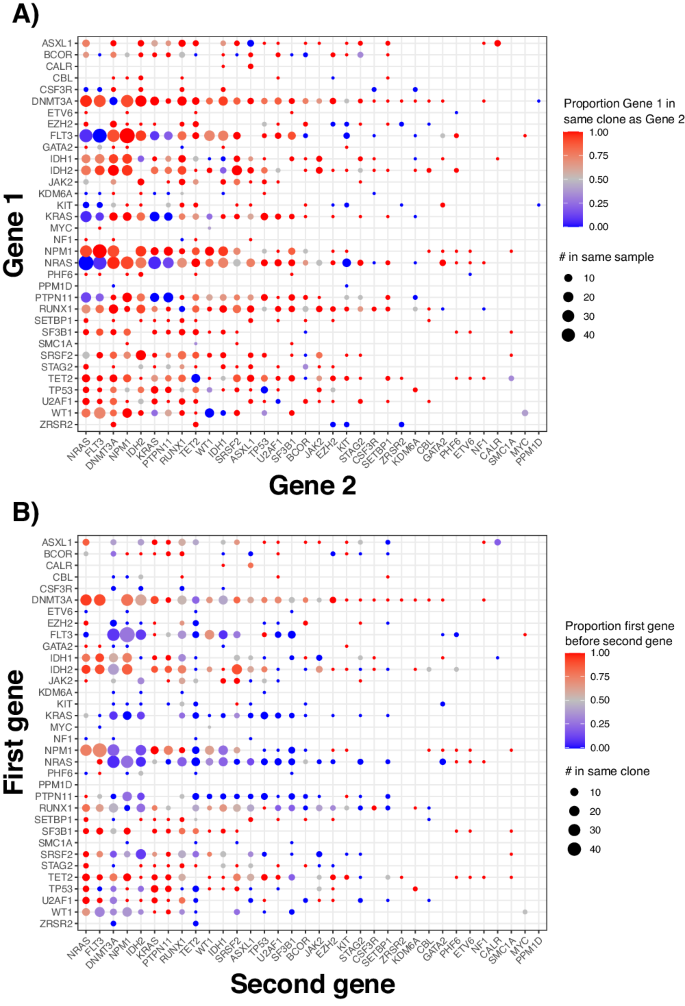
Utilizing SCITE (16,17), we created a mutation tree from every affected person’s mutation matrix (e.g., Fig. 1A). The bushes had variable numbers of pathways and genes per pathway (Fig. 1B, C), and the most typical pairwise hyperlinks between mutations concerned NPM1, DNMT3A, FLT3, NRAS, and IDH2 (Fig. 1D). These orderings had been corroborated by bulk sequencing since variations in variant allele frequency (VAF) from sequencing achieved utilizing the identical samples and variants correlated with variations within the mutated share of cells for pairs of variants in the identical clones (Pearson correlation 0.57, p = 2 × 10−51, Supplementary Fig. 7B). Amongst all bushes, 49% had branched evolution (48% when limiting to mutations from prognosis). Of the 101 bushes that had branched evolution, signaling mutations represented 66% of the occasions that instantly adopted a branching level (Supplementary Fig. 8A). In distinction, NPM1 mutations often served as a branching level (Supplementary Fig. 8B) as a result of NPM1 usually preceded signaling mutations; if branching occurred after an NPM1 mutation, 93% of such events concerned a signaling mutation vs. 28% if branching didn’t happen.
A Instance tree. Distributions of B the variety of distinct evolutionary pathways per tree (variety of bushes = 207), and C the typical variety of mutations per pathway. D Most typical two-gene evolutionary pathways mutated, when mutations had been summarized by gene. E All bushes merged, summarized by the gene by which the mutation is current, the place the dimensions of the node represents the variety of instances a specific pathway happens, ranging from the foundation node. Colours correspond to mutations, the place genes with related features have related colours (e.g., blue shades for DNA methylation and purple/orange shades for signaling mutations). F All bushes merged, the place the mutation occasions had been summarized by pathway and solely evolutionary pathways with at the least 5 occasions are depicted.
When summarizing mutations to genes (Fig. 1E), 224 distinct evolutionary orderings occurred throughout all sufferers (e.g., DNTM3A→NPM1 is vague from DNTM3A→NPM1→FLT3). Given the complexity of Fig. 1E, we merged bushes however summarized occasions based on the organic pathway corresponding to every gene (Fig. 1F, Supplementary Desk 2). Mutations associated to DNA methylation (e.g., DNMT3A, IDH1/2) had been often early, and terminal occasions had been usually signaling mutations. We additionally famous that DNA methylation mutations usually adopted different DNA methylation mutations, which had been pushed by particular kinds of DNA methylation mutations which can be much less related to AML development [23]. For instance, whereas DNMT3A R882, IDH1, and IDH2 mutations generally preceded NPM1 or signaling mutations (Supplementary Fig. 9A), DNMT3A non-R882 mutations often preceded different DNA methylation mutations (Supplementary Fig. 9B). Notably, DNMT3A R882 can be an instance the place sure mutations throughout the similar grouping (both the group of DNMT3A alone or the group of DNA methylation mutations) have completely different mutation orderings. Nevertheless, for ease of interpretation, we summarize mutation orderings on the gene and pathway ranges going ahead.
Pairwise mutation co-occurrence and order
To additional characterize the co-occurrence of mutations, we analyzed the frequency at which mutations occurred in the identical or completely different clones (Fig. 2A). Signaling mutations (Supplementary Desk 2) in the identical circumstances usually occurred in numerous clones. For example, completely different NRAS mutations occurred in distinct clones in 100% of circumstances. In distinction, NPM1 mutations almost at all times (>90% circumstances) co-occurred in the identical clone as mutations in signaling genes, DNA methylation genes, or transcription components (Fig. 2A).
A Plot exhibiting whether or not two mutations happen in the identical or completely different clone, summarized by gene. The scale of every dot represents the variety of instances mutations in two genes happen in the identical affected person pattern, and shade represents the frequency they’re in the identical clone. B Whether or not one mutation happens earlier than one other mutation. The scale of the dot represents the variety of instances they’re in the identical clone (not simply in the identical affected person pattern), and the colour represents the proportion of instances a mutation in a gene on the y-axis got here earlier than a mutation in a gene on the x-axis.
Many mutations additionally usually had attribute orderings relative to one another, akin to DNTM3A mutations occurring early and signaling mutations occurring late (Fig. 2B, Supplementary Desk 4), just like prior work [8]. Nevertheless, transcription components like RUNX1 and WT1 had variable mutation orderings, showing each earlier than and after mutations which can be usually early (e.g., DNMT3A) or late (e.g., FLT3).
Analyzing the order of mutation trios (relatively than pairs) corroborated these findings, the place trios usually started with DNA methylation mutations and terminated with signaling mutations (Supplementary Desk 5). Evolution of DNMT3A→NPM1→FLT3 was widespread, however different mutation trios had variable mutation orderings, like mixtures with DNA methylation and splicing mutations.
Between Supplementary Tables 4 and5, we additionally observe many different statistically important orderings. For instance, cohesin, splicing, and apoptosis-related (e.g., TP53) mutations usually happen after DNA methylation mutations however earlier than signaling mutations. Nevertheless, going ahead, we primarily deal with DNA methylation, NPM1, and signaling mutations as a result of these are mostly mutated on this dataset.
Unusual mutation orders
Though many mutation pairs occurred in attribute orders, we famous a number of circumstances the place mutation order deviated from typical patterns, akin to when signaling mutations occurred earlier than a DNA methylation or NPM1 mutation (Supplementary Fig. 10).
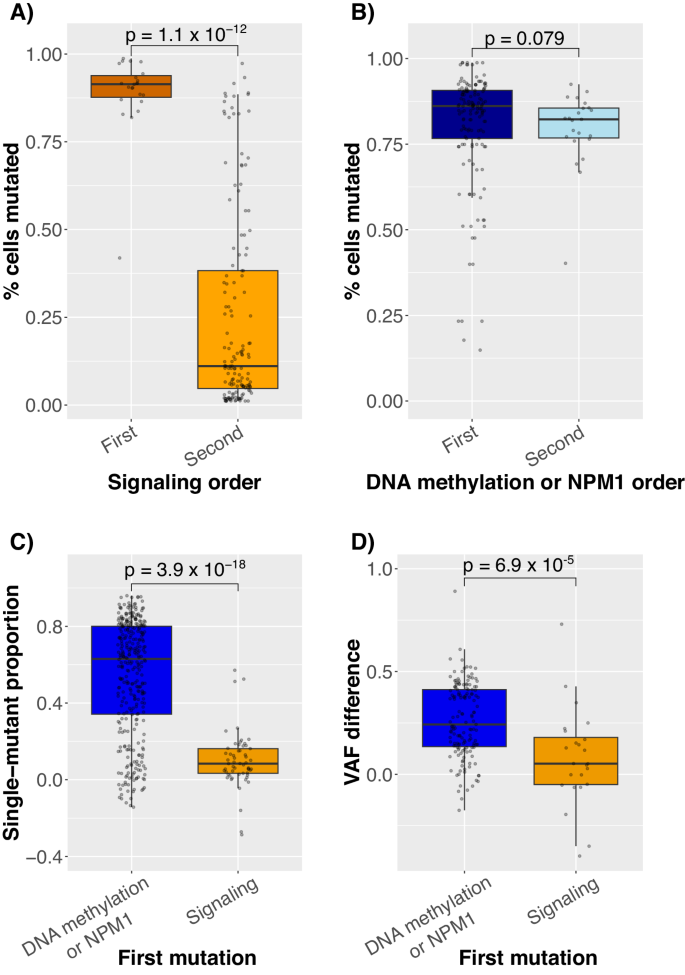
Earlier than characterizing these atypical orderings intimately, we validated their presence. First, if signaling mutations got here earlier than NPM1 or DNA methylation mutations, then the share of cells with these mutations must be larger. Certainly, the signaling mutation clone dimension in diagnostic samples was larger when the mutation got here earlier than (vs. after) the NPM1 or DNA methylation mutations (p = 1.1 × 10−12, Fig. 3A). Curiously, the share of cells with NPM1 or DNA methylation mutations was excessive regardless of relative signaling mutation order (Fig. 3B). Subsequent, if signaling mutations got here first, then each the share of mutated cells and the majority VAF must be larger than these of NPM1 and DNA methylation mutations. Certainly, throughout all samples and driver mutations, the signaling mutation’s share of mutated cells and VAF had been larger when it was first (89% [51/57] and 63% [15/24] of pairwise comparisons, respectively) and decrease when second (94% [318/338] and 93% [140/151]).
Boxplots exhibiting the A share of mutated cells containing a signaling mutation vs. whether or not the signaling mutation got here earlier than (First) or after (Second) an NPM1 or DNA methylation mutation. B Identical plot as (A) besides that the main focus is on the NPM1 or DNA methylation mutation p.c cells mutated. C Dimension of a single-mutant clone stratified by which mutation got here first. Single-mutant clone dimension was estimated by subtracting the proportion of cells with every mutation after eradicating cells the place there was no name for the mutation. This plot exhibits that the single-mutant clones for NPM1/DNA methylation-first circumstances had been larger than in signaling-first circumstances. D Distinction in variant allele frequency (VAF) utilizing bulk sequencing knowledge from the identical samples and variants. In A, B, solely diagnostic samples had been used for the reason that absolute quantity of illness might range with remedy, and the n = 148 for NPM1/DNA methylation-first and n = 23 for signaling-first. In C, D, for the reason that focus was on relative sizes of clones, all samples had been used, with n = 338 and n = 57 for NPM1/DNA methylation-first and signaling-first teams, respectively, and due to lacking bulk sequencing knowledge in (D), n = 151 and n = 24, respectively.
Though these outcomes corroborated the existence of signaling-first circumstances, the signaling mutation-only clones within the signaling-first circumstances had been constantly small. Utilizing the distinction within the share of mutated cells as a proxy for clone dimension, the single-mutant clone dimension was smaller in signaling-first circumstances than in NPM1/DNA methylation-first circumstances (p = 4 × 10−18, Fig. 3C). This distinction was additionally corroborated utilizing the distinction in bulk VAFs as a proxy for single-mutant clone dimension (p = 7 × 10−5, Fig. 3D).
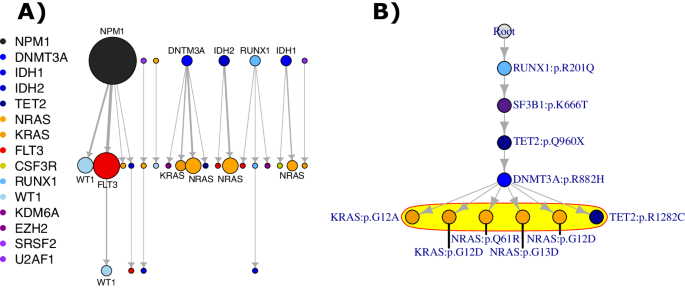
An identical sample of single-mutant clone dimension was beforehand seen in JAK2-first vs. TET2-first MPNs, the place JAK2-first circumstances had fewer single-mutant HSPCs, suggesting that TET2 mutation elevated the health of JAK2 mutation in HSPCs [6]. Thus, we suspected that NPM1 and DNA methylation mutations provided a selective benefit for signaling mutations amongst HSPCs in AML. We explored this phenomenon by inspecting new mutations throughout serial samples (25 prognosis/relapse pairs, 15 relapse/relapse pairs, 34 sufferers, Fig. 4A). Most new mutations at relapse had been signaling mutations (60%, 21/35), and new signaling mutations tended to come up after a beforehand current DNA methylation or NPM1 mutation. When contemplating all potential nodes in a tree from which signaling mutations may come up (together with the potential for no prior mutations), NPM1 and DNA methylation mutations disproportionately served because the speedy dad or mum node for a brand new signaling mutation (9/10 dad or mum nodes, Fisher’s check p = 0.002). For instance, in Fig. 4B, the NRAS mutations arose within the DNMT3A clone, regardless of the DNMT3A mutation being current in 41% of the sooner pattern’s cells in comparison with ≥90% of cells for the opposite mutations. As a result of signaling mutations disproportionately adopted DNA methylation and NPM1 mutations, NPM1 and DNA methylation m0utations might provide a bonus for signaling mutations in HSPCs.
A All new pathways at relapse throughout all out there paired serial samples within the single-cell dataset (derived from 25 prognosis/relapse and 15 relapse/relapse pairs, 34 sufferers complete). The highest layer of occasions represents occasions current within the prior pattern, though not essentially the preliminary occasion of a tree, and the decrease layers characterize occasions gained on a subsequent pattern. Genes with multiple occasion are labeled immediately. B Instance tree for which serial samples can be found, the place the occasions circled in yellow are new occasions on a subsequent pattern.
Scientific correlates with mutation order
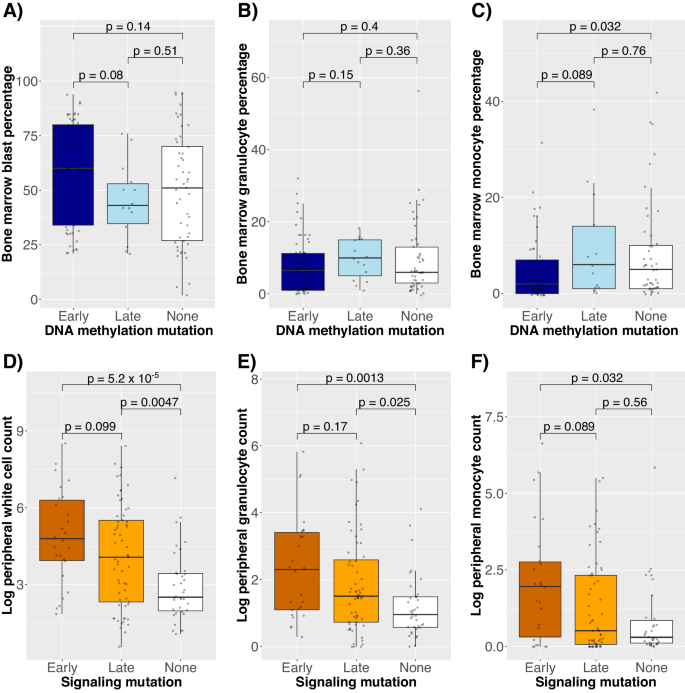
As a result of TET2 mutations change the HSPC steadiness in MPNs [6], we hypothesized that any benefit conferred by DNA methylation mutations in AML was partially because of the growth of extra immature HSPCs, obvious as blasts. To discover this, we in contrast “late” and “early mutations, that are those who happen with and with none previous mutations within the scDNAseq knowledge. Certainly, the bone marrow blast share was larger in diagnostic samples with early DNA methylation mutations in comparison with late DNA methylation mutations (p = 0.08, Fig. 5A), whereas the bone marrow granulocyte and monocyte percentages had been typically decrease (p = 0.15 and p = 0.09, respectively, Fig. 5B, C).
Distributions of A bone marrow blast share, B bone marrow granulocyte share, and C bone marrow monocyte share in comparison with whether or not a DNA methylation mutation was early, late, or not current within the pattern. Distributions of D log peripheral blast depend, E log peripheral granulocyte depend, and F log peripheral monocyte depend in comparison with whether or not a signaling mutation was early, late, or not current.
In distinction, signaling mutation order (see Supplementary Strategies for justification of the “early” and “late” categorization of signaling mutations) was not related to the bone marrow cell percentages (p ≥ 0.7 for all comparisons), but it surely was related to larger peripheral white blood cell (WBC) counts (p = 0.099, Fig. 5D). Though peripheral blast counts had been larger in signaling-early circumstances (median 14.8 vs. 3.7, rank-sum p = 0.14), so had been the peripheral granulocyte and monocyte counts (p = 0.17 and p = 0.089, respectively, Fig. 5E, F). Notably, we contemplate signaling mutations to be one group for easier interpretation, however they’ve completely different scientific phenotypes, akin to early NRAS/KRAS mutations having larger monocyte counts than later NRAS/KRAS mutations (p = 0.056), a pattern not seen for FLT3 mutations (p = 0.38).
To make sure that these associations between order and cell composition weren’t dataset-specific, we used proxies for early and late mutation order, particularly excessive and low VAFs (cutoff 0.3, beforehand used to outline dominant and clonal mutations [24, 25]), for validation within the BeatAML bulk DNA sequencing knowledge [22]. Early DNA methylation mutations had been certainly related to larger bone marrow blast percentages (p = 0.00041, Supplementary Fig. 11A). In distinction, whereas early signaling mutations weren’t related to bone marrow blast share (p = 0.35), they had been related to larger peripheral WBCs, granulocytes, and monocytes (p < 0.05 for all comparisons, Supplementary Fig. 11B–D).
Though these mutation orderings had distinct phenotypes, we additionally wished to tell apart whether or not the phenotype was associated to the order or the elevated clonal burden that resulted from a mutation occurring earlier. Thus, utilizing the scDNAseq knowledge, we carried out a number of linear regression adjusting for affected person age and the p.c of cells with the related mutation (Supplementary Desk 6). In multivariable analyses, DNA methylation clone dimension (p = 0.0079), however not mutation order (p = 0.21), was related to bone marrow blast share, suggesting that clone dimension mediated the affiliation between DNA methylation order and blast share (Supplementary Desk 6A). In an identical regression, signaling mutation clone dimension, relatively than mutation order, was considerably related to peripheral blast share (p = 0.0084, Supplementary Desk 6B). Nevertheless, signaling mutation order was independently related to peripheral granulocyte and monocyte counts (p = 0.088 and 0.035, respectively, Supplementary Desk 6B), suggesting that the order of signaling mutations, not simply the clonal burden, contributed to extra mature myeloid cell counts.
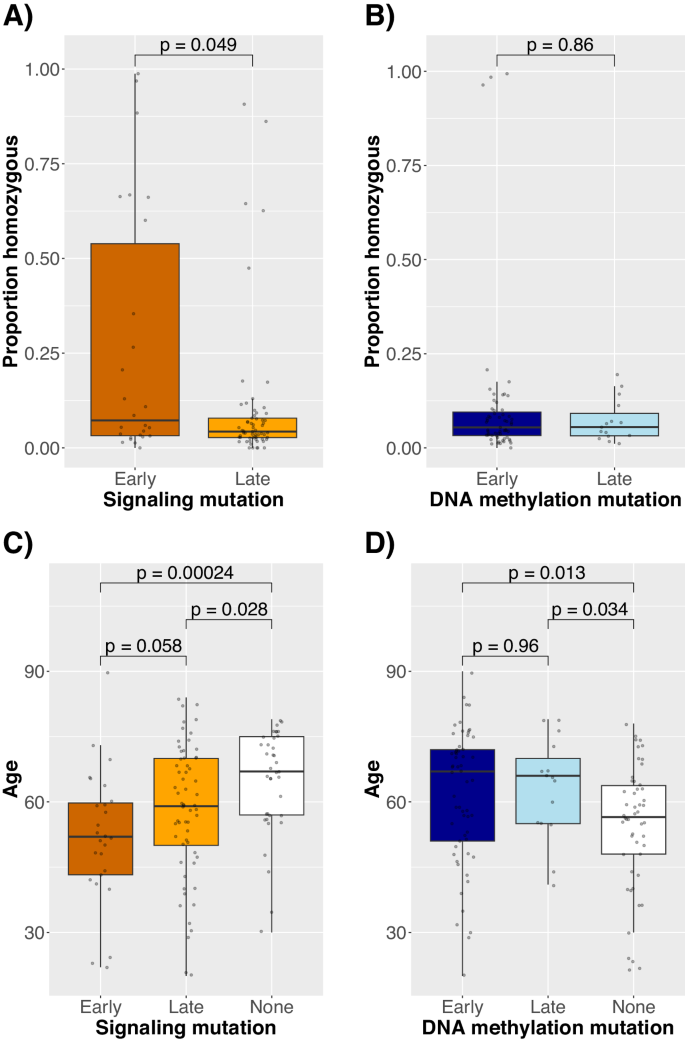
We subsequent examined whether or not mutation orderings in AML may clarify different affected person and illness traits, akin to youthful age and growing signaling mutation homozygosity, that are related to JAK2-first MPN circumstances [6]. Certainly, in diagnostic samples with early signaling mutations, signaling mutations had been extra usually homozygous (median 5% vs. 21% of cells homozygous, p = 0.049, Fig. 6A), and sufferers had been youthful (median 52 vs. 59 years outdated, p = 0.058, Fig. 6C). In distinction, the identical patterns didn’t maintain for DNA methylation mutations (Fig. 6B, D). Notably, the affiliation with signaling mutation homozygosity was pushed by a minority of circumstances (Fig. 6A) and primarily FLT3 (p = 0.011), for which lack of heterozygosity has beforehand been related to poor prognosis [26]. Though detecting zygosity in scDNAseq knowledge might be confounded by allele dropout, we discovered no proof of this since FLT3 mutation homozygosity was additionally not correlated with the variety of cells lacking mutation requires the related mutation or with sample-level allele dropout (Spearman correlation 0.04 [p = 0.78] and 0.07 [p = 0.65], respectively).
Signaling mutation (A, B) zygosity and affected person age (C, D) at prognosis in comparison with whether or not signaling (A, C) and DNA methylation (B, D) mutations had been early or late (or there was no mutation, within the age comparability) amongst diagnostic samples. “Early” signifies that no mutations are recognized to happen earlier than it based mostly on the scDNAseq dataset.
This constellation of evolutionary patterns and scientific correlates involving signaling mutations additionally creates the potential to higher perceive different mutations. For instance, WT1 mutations contribute to relapse [27] however have an unclear position in AML pathogenesis [28], and we discovered that WT1 mutations share many traits with signaling mutations. Like mutations in FLT3 and NRAS, WT1 mutations often occurred in NPM1-mutant clones (Figs. 2A and 4A); early WT1 mutations usually occurred in youthful sufferers; and WT1-first circumstances had small single-mutant clones when co-occurring with NPM1 mutations (Supplementary Fig. 12). In multivariable analyses, early WT1 mutations had been additionally related to age and better neutrophil and monocyte counts (Supplementary Desk 6C).
Though we discovered a number of phenotype variations related to mutation order between DNA methylation and signaling mutations, sufferers with these completely different orderings didn’t have considerably completely different total survival (Cox regression age-adjusted p = 1 for signaling vs. DNA methylation first). Amongst comparatively prevalent mutation orderings, SF3B1→FLT3 was almost considerably related to a worse prognosis after false discovery charge correction (age-adjusted hazard ratio 5.6, q worth = 0.056, Supplementary Fig. 13A). Nevertheless, this affiliation was not important after adjusting for the presence of an SF3B1 mutation (p = 0.44), which itself carries a poor prognosis [4].
Nonetheless, exploratory analyses of different phenotypes at prognosis (Supplementary Fig. 13B–E) revealed significant associations, akin to evolution involving IDH1/IDH2 mutations and decrease granulocyte (median 1.7 vs. 3.0, p = 3.6 × 10−6) and monocyte counts (median 1.2 vs. 1.9, p = 0.0063), or orderings with SRSF2 occurring predominantly in older people (median age 73 vs. 59, p = 0.017).