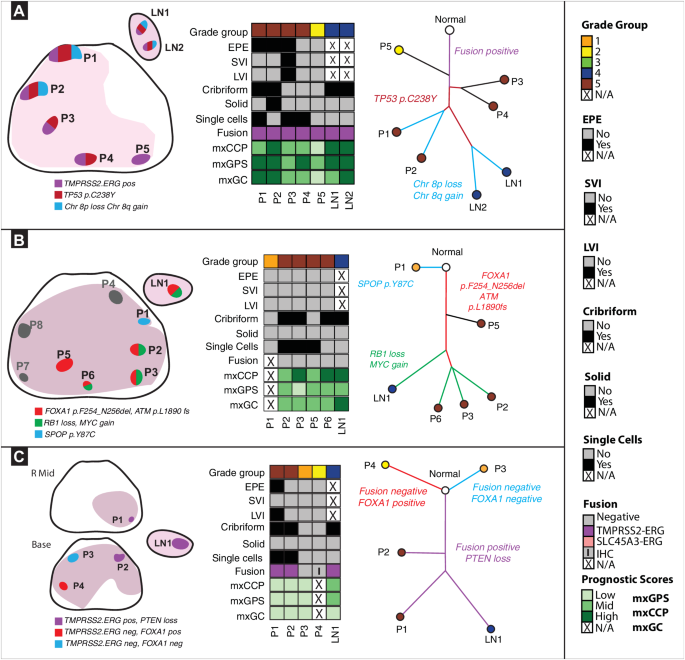
Mapping major prostate most cancers to synchronous LN metastasis
From a cohort of 18 sufferers (age 50–73 years) who had radical prostatectomy and pelvic lymph node (LN) dissection for prostate most cancers, we carried out multi-region sampling from formalin fastened paraffin embedded (FFPE) blocks yielding 103 major tumor and 28 LN metastasis samples (affected person knowledge is summarized in supplementary file – Supply Knowledge). We carried out excessive depth, focused, multiplex DNA sequencing to characterize the genomic profile of every tumor area utilizing two focused DNA NGS panels: the Complete Most cancers Panel (CCP; 409 genes and 15,992 amplicons) and a customized Pan-GenitoUrinary (Pan-GU) most cancers panel (135 genes and 3127 amplicons). Focused RNA NGS sequencing was additionally carried out utilizing a customized prostate cancer-focused NGS panel to guage the gene fusion standing of every pattern and derive related tissue-based prognostic scores [Myriad Prolaris™ Cell Cycle Progression (mxCCP) score, Oncotype DX™ Genomic Prostate Score (mxGPS), and Decipher™ Genomic Classifier (mxGC)] as beforehand carried out7 (Fig. S1). We decided and in contrast histopathological traits, RNA tissue-based prognostic signatures, somatic DNA mutations, copy quantity alterations (CNA), and gene fusion standing between major and LN illness (Fig. 1).
We used two focused DNAseq panels to establish key somatic mutations and replica quantity alterations (CNA) throughout 10 sufferers who handed our customized high quality management filtering standards. For CNA evaluation, solely the highest 445 genes (no. of amplicons per gene > 4 & log10 false discovery charge <0.01 & absolute log2CNvalue > 0.3) with losses and features are displayed within the heatmap. Unsupervised hierarchical clustering of all tumor areas inside every affected person was carried out to interrogate major tumor areas that cluster with their respective synchronous LN metastasis areas utilizing log2 normalized knowledge. Genes have been ordered by the chromosome quantity together with their begin and finish positions inside every chromosome. CNA for the identified prostate cancer-relevant genes are annotated. ETS gene fusion standing was derived from focused RNAseq knowledge utilizing an in-house fusion quantification pipeline. Related clinicopathologic variables equivalent to grade, stage, extraprostatic extension (EPE), seminal vesicle invasion (SVI), lymphovascular invasion (LVI), cribriform sample, stable sample, single cells and derived commercially accessible prognostic scores (mxCCP (derived Cell Cycle Development rating or ProlarisTM), mxGPS (derived Genomic Prostate Rating or OncotypeTM), and mxGC (derived Genomic Classifier or DecipherTM) for every pattern are annotated on the heatmap. Prognostic scores have been categorized into low, mid, and excessive teams primarily based on their Q1 and Q3 values for comparability amongst samples with the identical affected person in addition to comparability throughout totally different scores inside every pattern. We noticed intra- and inter-patient heterogeneity in histologic grade, genomic alterations, and derived prognostic gene signatures. Supply knowledge are supplied as a Supply Knowledge file.
After high quality management (QC) steps as described within the strategies, 10 sufferers (65 major tumor and 16 LN metastatic samples) had enough knowledge for phylogenetic analyses. All sufferers within the analytic cohort exhibited adversarial pathological traits, together with cribriform, single cell, or stable cell sample in not less than one tumor focus. A complete of eight sufferers had EPE, one demonstrated SVI and 4 confirmed proof of lymphovascular invasion (LVI). We famous substantial transcriptomic heterogeneity, together with discordant-derived prognostic gene signature scores each inside lesions and between lesions in the identical affected person (Fig. 1). Recurrent CNAs have been assessed from focused NGS knowledge utilizing an strategy that was beforehand benchmarked to fluorescence in situ hybridization (FISH), comparative genomic hybridization (CGH) array, and entire exome sequencing (WES) knowledge, exhibiting a excessive diploma of concordance amongst these approaches23. In a subset of samples in our cohort (n = 8 samples; from affected person #1), we additionally carried out low-pass entire genome sequencing (LPWGS) and located excessive concordance with recurrent CNAs recognized utilizing focused DNA NGS equivalent to 8p loss and 8q achieve (Fig. S2). TMPRSS2:ERG fusion transcripts have been recognized in 4 sufferers and the SLC45A3:ERG fusion transcripts have been detected in a single affected person (Fig. 1, Fig. S1). Phylogenetic analyses have been carried out with R phangorn bundle utilizing neighbor becoming a member of methodology to computationally reconstruct the clonal evolution of prostate most cancers and decide the seemingly clonal origin of LN metastasis for every affected person (Fig. 2, S3–10)19,24. Moreover, we utilized PhyloWGS as an orthogonal methodology to generate phylogenetic reconstructions in a subset of sufferers (n = 3) and noticed typically concordant outcomes (Fig. S3 and Fig. 2). Our knowledge reveal a number of potential histopathologic and molecular components related to illness unfold as described beneath.
Supply knowledge are supplied as a Supply Knowledge file. A. Spatial unfold linked to extra-prostatic extension (EPE). Left panel: In affected person #1, 5 major and a couple of LN areas encompassing Grade Teams (GG) 2-5 have been analyzed. We recognized three distinct molecular subtypes. Center panel: All tumor areas, together with the 2 LN metastases areas, have been TMPRSS2:ERG fusion-positive and with heterogeneous prognostic gene signatures. Proper panel: TP53 somatic mutation was detected on all tumor areas, apart from P5. Key chromosomal alterations, equivalent to 8p loss and 8q achieve, have been solely detected in areas P1, P2, LN1, LN2. Right here, the information recommend that major tumor areas P1, P2 carefully resemble LN1, LN2. Moreover, P5 represents a definite subclone unrelated to different major areas or LNs. B. Driver alterations in metastasis to LN. Left panel: In affected person #33, 8 major tumor (GG1-5) and 1 LN metastasis area have been analyzed. Grey areas (P4,7,8) designate these with low tumor purity that have been excluded. We recognized three molecular subtypes. Center panel: All areas have been ETS fusion destructive apart from P1, P8. P4, P7, and P8 areas have been excluded as a consequence of low tumor content material. Proper panel: An SPOP mutation was detected solely in P1. Driver alterations: FOXA1, ATM frameshift mutations have been detected in areas P2, P3, P5, P6, LN1. RB1 loss and MYC achieve have been seen in P2, P3, P6, and LN1, however not P5. Right here, knowledge recommend that major areas P2, P3, P6, all GG5 areas with cribriform patterns, carefully resemble LN1. C. LN metastasis in multiclonal major illness. Left panel: In affected person #41, 4 major (GG2-5) and 1 LN metastasis area have been analyzed. P1, a area of EPE, was taken from the mid prostate and P2-P4 was taken prostate base. We recognized three distinct tumor clones. Center panel: All samples besides P3 and P4 have been TMPRSS2:ERG fusion-positive. Proper panel: P4 had a FOXA1 somatic mutation which was not seen in different areas. Right here, knowledge recommend that major area P1 carefully resembles LN1, as each are TMPRSS2:ERG fusion-positive and harbor PTEN loss.
Metastatic unfold of prostate most cancers is linked to aggressive pathologic options
A number of pathologic options have been related to LN metastasis, together with histologic grade, cribriform or single cell sample, and EPE (Fig. 1). For instance, all 10 sufferers had not less than one major tumor area with a cribriform sample, and cribriform sample was noticed in each the dominant major tumor area and LN metastasis in seven sufferers. Apparently, a single cell sample – related to excessive histologic grade – was detected in LN specimens in solely two sufferers (20%), although it was current within the major tumors of eight sufferers. Intriguingly, in 8 sufferers with EPE, phylogenetic reconstruction supported the area of EPE because the seemingly supply of the LN metastasis in 4 instances (Fig. 2a, S4,7,8). In affected person #1 (Fig. 2a and S3a), all tumors harbored a TMPRSS2–ERG gene fusion. Whereas 4 major tumor areas confirmed concordant TP53, IL6ST, and TPR mutations, solely two major tumor areas (P1 and P2, each EPE) additionally harbored an LRP1B mutation and high-level CNAs (e.g., like 16q loss and 8p12 loss) that have been additionally current within the two LN (LN1 and LN2) metastasis foci. Notably, major tumor areas P3 and P4, with the presence of single cells, didn’t seem to have seeded the LN metastatic foci. On this affected person, major tumor areas P1 and P2 (GG5 areas with EPE) have been most probably the supply of LN metastasis. In affected person #2 (Fig. S4), all tumor areas have been ETS gene fusion destructive. A CDK12 frameshift mutation with MYC and FGFR3 achieve was additionally detected in all areas. Phylogenetic evaluation suggests P4 (a spotlight with EPE) most carefully resembles the LN1 metastasis. In affected person #34 (Fig. S7), BRCA1 and PTEN losses have been seen in P1, P4, P7, P8, P9, LN1, LN2, and LN3, however not in P2, P3, and P5, suggesting two totally different branches of clonal evolution. Areas P4 and P8 confirmed proof of EPE. Phylogenetic evaluation means that areas P1, P4, P7, P8, and P9 are most probably the supply of LN metastasis on this affected person. In affected person #38 (Fig. S8), areas P3, P4, P5, P6, and P7 (a area of EPE) displayed FOXA1 mutations together with losses of PTEN and RB1, much like the LN foci (LN1). The world of P6 and LN1 shared extra lack of CDKN2B and achieve of RECQL4, suggesting this because the most probably clonal supply of the LN metastatic focus, although the area with EPE (P7) seemingly contributed given its shared mutational burden and proximity to P6 and LN1 by phylogenetic evaluation. Taken collectively, these findings recommend that aggressive pathologic options – EPE, histologic grade, and/or cribriform sample – could also be related to the event of LN metastases.
Position of driver alterations and genomic complexity in prostate most cancers metastasis to LNs
As proven in Fig. 1, established early oncogenic driver alterations together with ERG gene fusions in addition to FOXA1, CDK12, and SPOP mutations have been usually current in each the first tumor and LN metastatic foci. The presence of a number of such alterations inside three sufferers in our cohort (Fig. 2 and S7) suggests the opportunity of tumor multiclonality – a well-established idea in prostate most cancers5,6,9. Importantly, in these sufferers, NGS analyses solely confirmed proof of a single early oncogenic driver within the LN metastases, supporting the monoclonal principle of metastatic prostate most cancers (see beneath). Moreover, our analyses revealed substantial intratumoral heterogeneity throughout the sampled major tumor areas, together with TP53 mutations and genomic complexity. Intriguingly, though TP53 mutations have been recognized in a number of major tumor areas from two sufferers (#1 and #40, Fig. 2 and S10), this mutation was solely noticed within the LN metastatic foci from affected person #1. Conversely, TP53 mutations have been detected solely within the LN metastatic foci from two different sufferers (#4 and #30, Fig. S5 and S6). Regardless, these knowledge recommend that TP53 mutations could also be related to metastatic development of major prostate most cancers.
Elevated genomic complexity was additionally regularly noticed in LN metastatic foci. For instance, in a affected person with organ-confined (pT2) prostate most cancers and LN metastasis (affected person #33, Fig. 2b and S3b), the samples demonstrated genomic complexity with FOXA1 and ATM frameshift mutations detected in some major tumor and LN metastasis areas (P2, P3, P5, P6, LN1); an SPOP mutation was detected solely in a GG1 major tumor focus (P1); and RB1 loss and MYC achieve have been shared between the presumed dominant major tumor areas and the LN metastasis focus (P2, P3, P6, LN1). In affected person, #41 (Fig. 2c), areas P1, P2 and LN1 all have been TMPRSS2:ERG gene fusion-positive and harbored PTEN loss. P1 most carefully resembled LN1 primarily based on phylogenetic evaluation, suggesting this because the area that seemingly gave rise to the LN1 metastatic focus. Within the PhyloWGS strategy, nevertheless, (Fig. S3c), P2 was recognized as carefully resembling the area of the LN metastasis (LN1). Notably, on this strategy, P1 was filtered out by the algorithm regardless of having the TLX1 mutation as a result of a clonal lack of 8q was not discovered on this pattern. Nevertheless, it’s noteworthy that areas P1 and P2 look like spatially associated anatomically (Fig. 2c, left panel). In affected person #2 (Fig. S4), LN1 and P4 shared a CDKN2B loss, together with a CDK12 frameshift mutation and MYC and FGFR3 loss as mentioned above. In affected person #30 (Fig. S6), we noticed shared lack of CDKN2A, ERCC2, and ERCC3 in lesions P1, P3, P4, P7 and LN2. LN2 had a further mutation in TP53. Of all of the tumor areas, P3 most resembled LN2 in mutational profile and genomic complexity, with phylogenetic evaluation suggesting this because the seemingly clonal origin of LN2 metastasis. In affected person #34 (Fig. S7), PTEN and BRCA1 losses have been noticed amongst P1, P4, P7, P8, P9, and LN1, LN2, and LN3. In affected person #38 (Fig. S8), P6 and LN1 each displayed CDKN2B loss and RECQL4 achieve as mentioned above. Lastly, affected person #4 (Fig. S5) confirmed important genomic complexity and extra focused NGS with a 500-gene panel revealed an MUTYH mutation with tumor mutational burden and microsatellite instability in areas P1 and P2, suggesting these areas developed continued genomic aberrations after LN1 and LN2 metastasis (as advised by earlier clonal branching of the LN metastasis on phylogenetic tree).
Taken collectively, these knowledge assist the potential position of driver genomic alterations within the growth of synchronous LN metastasis in major prostate most cancers.
Lymph node metastatic homogeneity
We noticed histologic, genomic, and transcriptomic heterogeneity throughout the first prostate most cancers foci (Fig. 1, S1). A complete of 5 sufferers had a number of LN areas analyzed. These synchronous LN metastases have been usually homogenous inside a given affected person, in keeping with present literature relating to metastatic foci19,20. Notably, a number of histopathologic options have been related to LN metastasis, together with GG and cribriform or single cell sample (Fig. 1). In sufferers with a number of LN foci analyzed, we famous principally homogenous patterns of mutations, suggesting a shared clonal origin. For instance, in affected person #1 (Fig. 2a and S3), each LN1 and LN2 shared chr8p loss and chr8q achieve, in addition to mutations in TP53. In affected person #4 (Fig. S5), LN1 and LN2 shared lack of CDKN2B, FANCA achieve, and TP53 frameshift mutations. All three LN metastatic areas LN1, LN2, and LN3 in affected person #34 (Fig. S7) shared PTEN, BRCA1, BRCA2, and CDKN2A losses in addition to MYC achieve.
In distinction, two of the 5 sufferers with a number of LN foci analyzed displayed discordant molecular profiles and thus separate branching within the phylogenetic evaluation, together with affected person #2 (Fig. S4) with LN1 and LN2 exhibiting discordant losses of CDKN2B (LN1) and TP53 (LN2). In affected person #30 (Fig. S6), LN2 shared options of CDKN2A, ERCC2, ERCC3 loss, and TP53 mutations, whereas LN1 didn’t present these shared alterations. Whereas low genomic complexity, early clonal branching or evolution, or low tumor content material are potential explanations for these observations, the opportunity of LN metastasis from separate tumor areas or clones can’t be excluded.