Identification of APA-related LUAD genes
A complete of 518 APA-related genes in LUAD ( | Rs | > 0.3, PFDR < 0.05) had been obtained. Amongst these, the PDUI values of 285 genes had been positively correlated with the corresponding gene expression (Rs > 0.3, PFDR < 0.05), and the PDUI values of the opposite 233 genes had been negatively correlated with the corresponding gene expression (Rs < -0.3, PFDR < 0.05).
We additional analyzed the expression variations of the above 518 genes at mRNA degree between 57 paired LUAD tumor tissues and adjoining non-tumor tissues utilizing the TCGA database. The consequence confirmed {that a} whole of 143 genes (mRNA degree) had been differentially expressed ( | FC | > 1.5, P < 0.05). Then, we validated these 143 genes between 49 paired LUAD tumor tissues and adjoining non-tumor tissues from the Chinese language inhabitants, and 65 genes had been validated as considerably differential expressed ( | FC | > 1.5, P < 0.05).
Primarily based on these validated 65 genes on the mRNA degree, we additional evaluated whether or not they had been differentially expressed on the degree of their corresponding coding proteins.
The outcomes confirmed that there have been 32 proteins differentially expressed between the tumor tissues and adjoining non-tumor tissues from the identical Chinese language inhabitants and the expression instructions of 32 proteins had been per the corresponding mRNAs.
Thus, the overlapped 32 APA-related LUAD genes with constant differential expression each on the mRNA and its′ coding protein ranges had been used for additional examine.
Identification of apaQTL/eQTL-SNPs in APA-related LUAD genes
A complete of 423 apaQTL-SNPs positioned within the above 32 gene areas (PFDR < 0.05) had been obtained via a public 3′aQTL-atlas web site, whereas 338 apaQTL-SNPs additionally confirmed eQTL features and should affect expression ranges of 9 APA-related LUAD genes (P < 0.05). Subsequently, 256 apaQTL/eQTL-SNPs had been survived with MAF > 0.05 in CHB. Lastly, 28 apaQTL/eQTL-SNPs had been chosen after LD filter (r2 < 0.8) (Fig. 1) and their detailed info was proven in Desk 1.
The affiliation between candidate apaQTL/eQTL-SNPs and LUAD threat
We additional examined the affiliation between 28 apaQTL/eQTL-SNPs and LUAD threat within the Chinese language inhabitants utilizing the China Nanjing Lung Most cancers GWAS database. As proven in Desk 2, the variant T allele of rs10452178 positioned in CISD2 was considerably related to a decreased threat of LUAD (OR = 0.92, 95% CI = 0.87–0.98, P = 0.009). The variant T allele of rs277646 positioned in NIT2 was considerably related to an elevated threat of LUAD (OR = 1.12, 95% CI = 1.02–1.22, P = 0.015). Apart from, the variant T allele of rs11714045 positioned in NIT2 confirmed a borderline vital affiliation with an elevated threat of LUAD (OR = 1.05, 95% CI = 1.00–1.10, P = 0.076).
The correlation between PDUI worth and corresponding gene expression
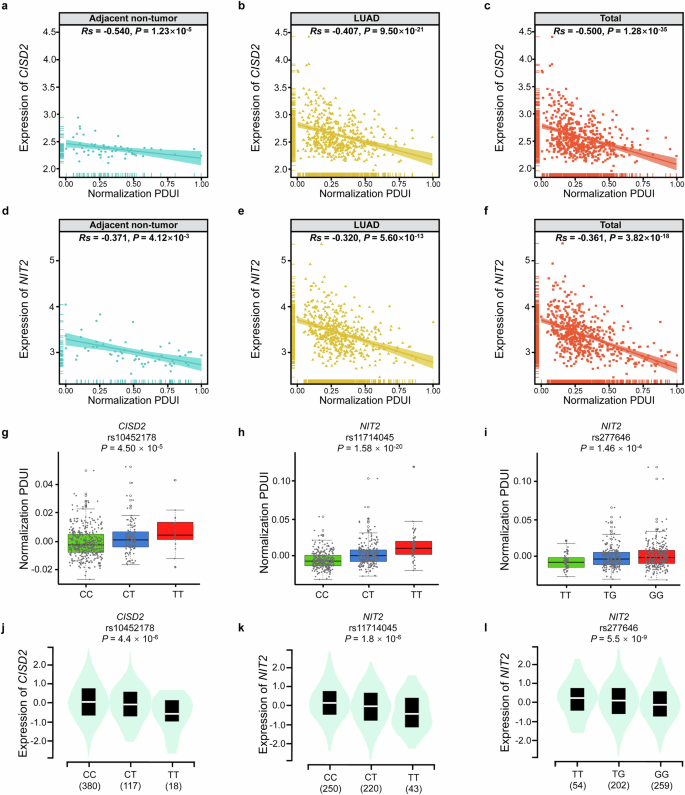
To research the APA utilization of CISD2 and NIT2 in LUAD, we mixed PDUI knowledge and gene expression knowledge from TCGA. As proven in Fig. 2a–c, in adjoining non-tumor tissues (Rs = -0.540, P = 1.23 × 10−5), LUAD tumor tissues (Rs = -0.407, P = 9.50 × 10−21), and whole tissues (Rs = -0.500, P = 1.28 × 10−35), all of us noticed a damaging correlation between the PDUI worth and gene expression degree of CISD2. Because the PDUI worth elevated, the expression of CISD2 decreased. As well as, the PDUI worth of NIT2 was additionally negatively correlated with NIT2 gene expression degree in each adjoining non-tumor tissues (Rs = −0.371, P = 4.12 × 10−3), LUAD tumor tissues (Rs = −0.320, P = 5.60 × 10−13), and whole tissues (Rs = -0.361, P = 3.82 × 10−18) (Fig. 2nd–f).
a–c The correlation between PDUI worth and CISD2 expression in adjoining non-tumor tissues (a), LUAD tumor tissues (b) and whole tissues (c). d–f The correlation between PDUI worth and NIT2 expression in adjoining non-tumor tissues (d), LUAD tumor tissues (e) and whole tissues (f). g–i The connection between totally different genotypes of rs10452178 and CISD2 PDUI values (g), rs11714045 and NIT2 PDUI values (h), rs277646 and NIT2 PDUI values(i). j–l The connection between totally different genotypes of rs10452178 and CISD2 expression (j), rs11714045 and NIT2 expression (okay), rs277646 and NIT2 expression (l).
APA evaluation of three recognized apaQTL/eQTL-SNPs
As proven within the Fig. 2g–i, two apaQTL/eQTL-SNPs (rs10452178 and rs11714045) had considerably greater PDUI values beneath the variant alleles (P = 4.50 × 10−5, P = 1.58 × 10−20), whereas the opposite apaQTL/eQTL-SNP (rs277646) had considerably decrease PDUI values beneath the variant T allele (P = 1.46 × 10−4).
eQTL evaluation of three apaQTL/eQTL-SNPs
The eQTL evaluation indicated considerably decrease expression ranges of CISD2 beneath the variant T alleles of rs10452178 (P = 4.4 × 10−6), and equally, considerably decrease expression ranges of NIT2 was noticed beneath the variant T alleles of rs11714045 (P = 1.8 × 10−6). Apart from, considerably greater expression ranges of NIT2 was noticed beneath the variant T alleles of rs277646 (P = 5.5 × 10−9) (Fig. 2j–l).
Expression evaluation of CISD2 and NIT2
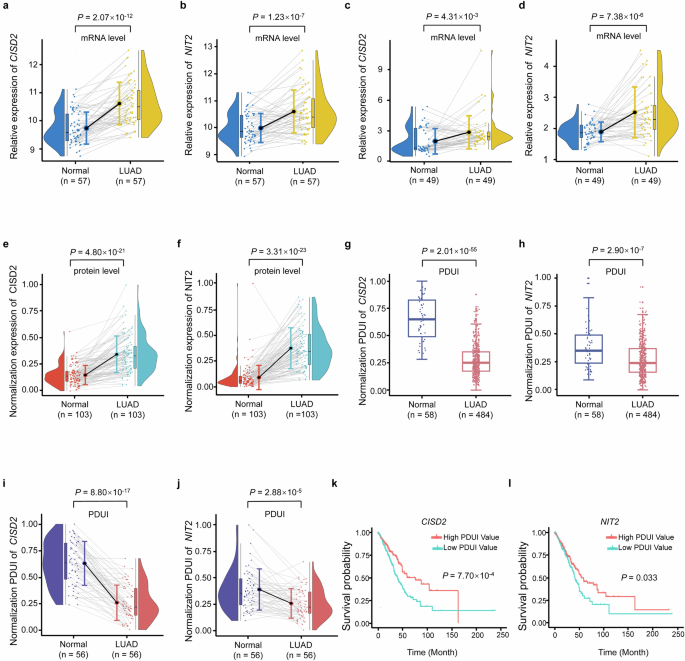
The mRNA expression degree of CISD2 (P = 2.07 × 10−12) (Fig. 3a) and NIT2 (P = 1.23 × 10−7) (Fig. 3b) had been each considerably greater within the LUAD tumor tissues (n = 57) from TCGA database. We additional validated the mRNA expression degree of CISD2 and NIT2 within the Chinese language inhabitants. The consequence confirmed that the mRNA expression degree of CISD2 and NIT2 had been additionally greater in LUAD tumor tissues (n = 49) in contrast with paired adjoining non-tumor tissues (P = 4.31 × 10−3, P = 7.38 × 10−6) (Fig. 3c, d). As well as, their corresponding coding proteins additionally confirmed the identical development (P = 4.80 × 10−21, P = 3.31 × 10−23), which had been up-regulated in LUAD tumor tissues (Fig. 3e, f).
a, b CISD2 mRNA expression (a) and NIT2 mRNA expression (b) within the LUAD tumor tissues and paired adjoining non-tumor tissues within the TCGA dataset. c, d CISD2 mRNA expression (c) and NIT2 mRNA expression (d) within the LUAD tumor tissues and paired adjoining non-tumor tissues within the Chinese language inhabitants. e, f CISD2 protein expression (e) and NIT2 protein expression (f) within the LUAD tumor tissues and paired adjoining non-tumor tissues within the Chinese language inhabitants. g, h The PDUI worth of CISD2 (g) and NIT2 (h) between LUAD tumor tissues and adjoining non-tumor tissues. i, j The PDUI worth of CISD2 (i) and NIT2 (j) between LUAD tumor tissues and paired adjoining non-tumor tissues. okay, l Survival evaluation of PDUI worth of CISD2 (okay) and NIT2 (l).
PDUI evaluation of CISD2 and NIT2
Utilizing TCGA database, the PDUI worth of CISD2 and NIT2 had been considerably decrease in whole LUAD tumor tissues in contrast with adjoining non-tumor tissues (P = 2.01 × 10−55, P = 2.90 × 10−7) (Fig. 3g, h). We additional carried out PDUI evaluation of CISD2 and NIT2 in 56 paired samples, and the outcomes additionally confirmed that the PDUI worth of CISD2 and NIT2 had been additionally considerably decrease in LUAD tumor tissues (P = 8.80 × 10−17, P = 2.88 × 10−5) (Fig. 3i, j). This means that the three′UTR size of CISD2 and NIT2 had been considerably shorter in LUAD tumor tissues in contrast with adjoining non-tumor tissues.
Survival evaluation of CISD2 and NIT2
To find out whether or not the incidence of APA occasions on this examine was associated to the survival of LUAD sufferers, we carried out survival analyses for sufferers from the TCGA dataset. The outcomes confirmed that the general LUAD survival is considerably longer in sufferers with excessive PDUI values of CISD2 and NIT2 than that with low PDUI values (P = 7.70 × 10−4, P = 0.033) (Fig. 3k, l).
The expression of CISD2 and NIT2 in cell traces
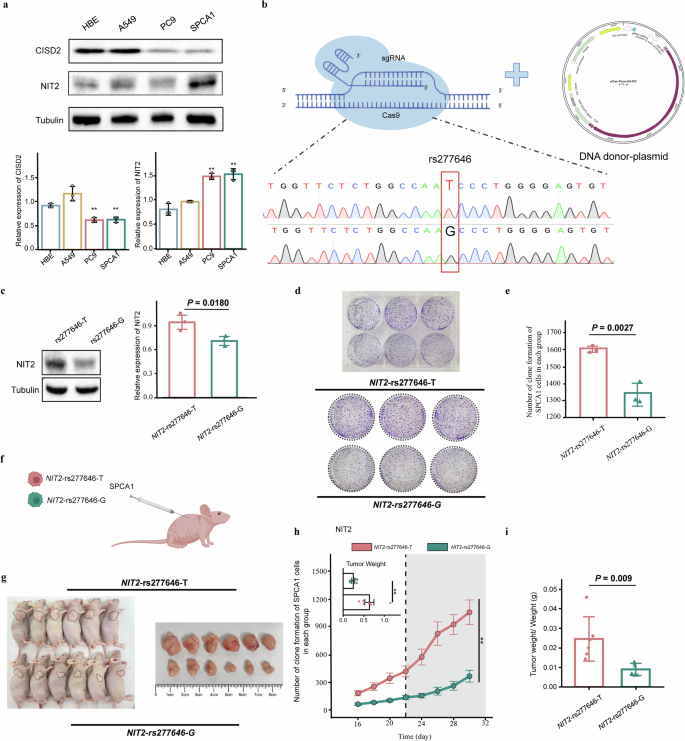
To find out the expression of CISD2 and NIT2 in LUAD cell traces, we carried out protein blotting on LUAD cell traces and regular HBE cell line. The outcomes point out that NIT2 expression in LUAD cell traces (PC9 and SPCA1) is considerably greater than in HBE cells. Moreover, there’s a development of upper expression of NIT2 within the LUAD cell line A549 in comparison with HBE cells. Nonetheless, the expression development of CISD2 in LUAD cell traces (A549, PC9, and SPCA1) just isn’t constant (Fig. 4a).
a The expression of CISD2 and NIT2 in cell traces. b NIT2-rs277646-G was obtained via genome enhancing mediated by CRISPR/Cas9. c The expression of NIT2 in NIT2-rs277646-T and NIT2-rs277646-G. d SPCA1 cell proliferation experiment after transfection. e The rs277646-T can promote the proliferation of SPCA1 cells. f Xenotransplantation animal mannequin. g Macroscopic statement of tumor nudity. h Comparability of tumor quantity modifications and weight between NIT2-rs277646-T group (n = 6 mice) and NIT2-rs277646-G group (n = 6 mice) in mice. i The ratio of tumor weight to physique weight in mice between the NIT2-rs277646-T group and the NIT2-rs277646-G group *P < 0.05, * * P < 0.01, * * * P < 0.001.
Results of NIT2-rs277646 on the malignant phenotype of LUAD in vitro and vivo
By CRISPR/Cas9 mediated genome enhancing, we efficiently obtained NIT2-rs277646-G and carried out DNA sequencing for validation (Fig. 4b). In the meantime, Western blot evaluation revealed that the expression of NIT2 within the NIT2-rs277646-T was greater than that within the NIT2-rs277646-G (P = 0.0180) (Fig. 4c). In contrast with the NIT2-rs277646-G group, there was a considerably greater cell proliferation capability within the NIT2-rs277646-T group (P = 0.0027) (Fig. 4d–e).
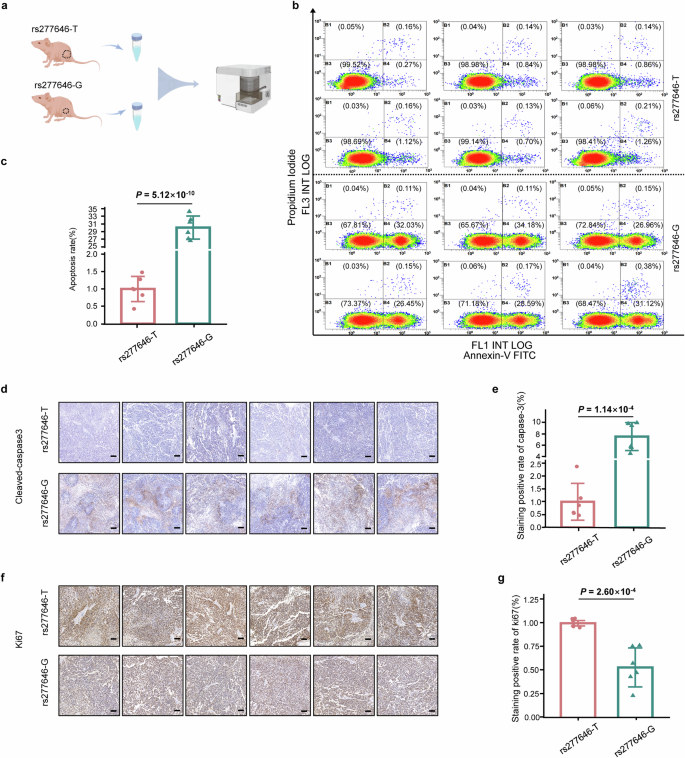
To find out the impact of various alleles of rs277646 on tumor development in vivo, SPCA1 cells transfected with NIT2-rs277646-T and NIT2-rs277646-G had been injected into BALB/c-nu mice to assemble an animal xenograft mannequin (Fig. 4f). The outcomes confirmed that the tumor quantity development price within the NIT2-rs277646-G group was slower, whereas the tumor quantity development price within the NIT2-rs277646-T group was sooner and all of the sudden elevated from the twenty second day (Fig. 4g, h). As well as, after killing the mice on the thirtieth day, the tumor weight of the NIT2-rs277646-T group considerably elevated in comparison with the NIT2-rs277646-G group (Fig. 4i). Move cytometry was carried out on tumor tissue samples from a mouse mannequin to detect and consider the apoptosis price of the SPCA1 cell line beneath T and G alleles of rs277646 (Fig. 5a). The outcomes confirmed that the apoptosis price of the NIT2-rs277646-T group was considerably decrease than that of the NIT2-rs277646-G group (Fig. 5b, c).
a Move cytometry sample diagram of major tumor cells extracted from mice. b, c Move cytometry was used to detect apoptosis of annexin V/PI. Harvest cells and use annexin V and 1 μ PI staining. Move cytometry confirmed that the cells had been within the phases of dwelling cells, early cell apoptosis, and late cell apoptosis. In contrast with the rs277646-G group, the apoptosis price of tumor cells beneath rs277646-T was lowered. d Picture of tumor slices from xenograft animals stained with Cleared Caspase-3 (100x goal). e The optimistic price of Cleared Caspase-3 within the rs277646-T group was considerably decrease than that within the rs277646-G group. f Picture of tumor slices from xenograft animals stained with ki67 (100x goal). g The optimistic price of ki67 staining within the rs277646-T group was considerably greater than that within the rs277646-G group.
Additional evaluation of mouse tumor tissue sections utilizing immunohistochemistry staining with cleaved-caspase 3 and ki67 confirmed that the optimistic price of caspase-3 staining within the rs277646-T group was considerably decrease than that within the rs277646-G group, whereas the optimistic price of ki67 staining within the rs277646-T group was considerably greater than that within the rs277646-G group (Fig. 5d–g), indicating the T allele of rs277646 could promote the malignant phenotype of LUAD.
Evaluation of poly(A) websites of NIT2 primarily based on the three′RACE experiment
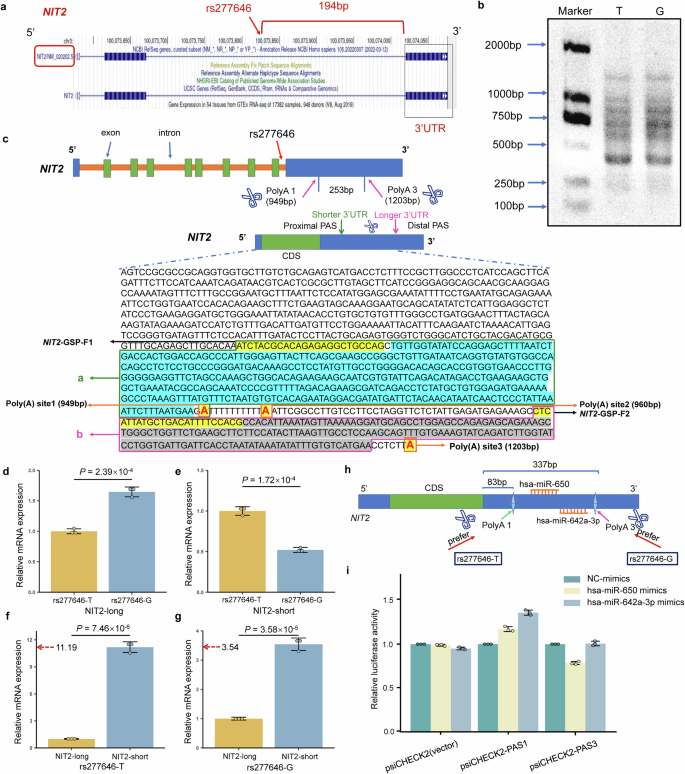
In line with the UCSC web site (https://genome.ucsc.edu/), rs277646 is positioned on the 194 bp upstream of the three′UTR of NIT2 (Fig. 6a). Subsequently, via the NCBI web site (https://www.ncbi.nlm.nih.gov/), it may be noticed that the size of NIT2 3′UTR is 6367 bp. On the identical time, there are 5 predicted poly (A) websites (PAS1-5) positioned at 949 bp, 961 bp, 1203 bp, 2472 bp, and 7233 bp, respectively. Subsequently, the three′RACE experiment confirmed that the NIT2 in SPCA1 cell line primarily selectively acknowledges the PAS1 and PAS3 websites (Fig. 6b, c).
a The connection between rs277646 on the NCBI web site and the three′UTR of the goal gene NIT2. b Agarose gel map of PCR merchandise of three′RACE beneath totally different alleles of rs277646. c Completely different alleles of rs277646 could mediate variations in poly (A) choice, leading to totally different 3′UTR transcript schematics and PCR product sequences. d The expression of the longer 3′UTR transcript of NIT2 within the rs277646-G allele is considerably greater than that within the T allele. e The expression of the shorter 3′UTR transcript of NIT2 within the rs277646-T allele is considerably greater than that within the G allele. f Within the pattern of rs277646-T allele, the expression of the shorter 3′UTR transcript of NIT2 was considerably greater than that of the longer 3′UTR transcript. g Within the pattern of rs277646-G allele, the expression of the shorter 3′UTR transcript of NIT2 was considerably greater than that of the longer 3′UTR transcript. h Sample Diagram of miRNA Binding to the NIT2 3′UTR Area. i Twin luciferase reporter gene assay for 3′UTR of NIT2 and miR-650, miR-642a-3p; n = 3. 3′RACE, 3′ fast amplification of cDNA ends; 3′UTR, 3′untranslated area; CDS, coding sequence; PAS, polyadenylation sign; GSP, gene-specific primer. *P < 0.05, * * P < 0.01, * * * P < 0.001.
The expression of NIT2 isoforms beneath totally different alleles of apaQTL/eQTL-SNP rs277646
The expression of the longer 3′UTR transcript of NIT2 was considerably decrease within the rs277646-T allele than that within the G allele (P = 2.39 × 10−4) (Fig. 6d), whereas the expression of the shorter 3′UTR transcript of NIT2 was considerably greater within the rs277646-T allele than that within the G allele (P = 1.72 × 10−4) (Fig. 6e). In the meantime, whether or not beneath the G allele or T allele of rs277646, the expression of the shorter 3′UTR transcript of NIT2 was considerably greater than that of the longer 3′UTR transcript (P = 7.46 × 10−6, P = 3.58 × 10−5) (Fig. 6f, g). On the identical time, the ratio of the shorter 3′UTR transcript to the longer 3′UTR transcript beneath the T allele of rs277646 (ratio=11.19) was greater than that beneath the G allele of rs277646 (ratio=3.54). This means that the NIT2 inclined to supply shorter 3′UTR transcripts beneath the T allele of rs277646.
The affect of hsa-miR-650 binding to the lengthy 3′UTR of NIT2
As a result of totally different genotypes of rs277646 affecting the expression of various subtypes of NIT2, we additional investigated its affect on gene-miRNA interactions. Firstly, we chosen microRNAs utilizing the ENCORI database (http://starbase.sysu.edu.cn/) and miRDB (https://mirdb.org/ontology.html), in the end figuring out hsa-miR-650 and hsa-miR-642a-3p as two microRNAs. In line with the prediction outcomes from the TargetScan web site (https://www.targetscan.org/vert_80/), each microRNAs are predicted to bind between Poly(A)1 and Poly(A)3. Particularly, Poly(A)1 is positioned 83 bp downstream from the beginning website of the NIT2 3′UTR, whereas Poly(A)3 is located 337 bp downstream from the beginning website of the NIT2 3′UTR. hsa-miR-650 binds to the area spanning 248-255 bp of the NIT2 3′UTR, whereas hsa-miR-642a-3p binds to the area spanning 320-326 bp of the NIT2 3′UTR (Fig. 6h). To validate the involvement of microRNAs on this interplay, we constructed quick sequences (~ PAS1) and lengthy sequences (~ PAS3) for the luciferase reporter gene assay.
Outcomes from the luciferase reporter gene assay confirmed that the hsa-miR-650 mimics or hsa-miR-642a-3p mimics didn’t bind to the quick 3’UTR of the NIT2 (~ PAS1). Nonetheless, within the lengthy 3’UTR of NIT2 (~ PAS3), the has-miR-650 mimics considerably lowered luciferase exercise, indicating that the has-miR-650 mimics binds to the lengthy 3’UTR of NIT2 (~ PAS3); The luciferase exercise of hsa-miR-642a-3p mimics didn’t lower, indicating that hsa-miR-642a-3p mimics doesn’t bind to the lengthy 3’UTR of NIT2 (~ PAS3) (Fig. 6i). Subsequently, the rs277646-T genotype results in NIT2 preferentially using the proximal poly(A) website, leading to shorter 3’UTR transcripts, which ends up in the lack of hsa-miR-650 binding websites on NIT2, thereby affecting the expression ranges of NIT2.