Drug tolerance is a dynamic fairly than a dormant state
Though an rising variety of research have targeted on the characterization of DTC and absolutely resistant proliferative cells (RPC)19,20,21,22,27,35,42, the kinetics of evolution by the totally different states is essentially unknown. We used the FUCCI (fluorescence ubiquitination cell cycle indicator) system43 to carry out real-time monitoring of the cell cycle dynamics in a panel of EGFR-mutant NSCLC cell traces handled with 1 µM EGFR-TKI (erlotinib or osimertinib). Importantly, cell traces have been beforehand subcloned to attenuate the presence of potential pre-existing resistant cells19,44 (Supplementary Fig. 1A–B). For all cell traces, we noticed a standard sample of G1 accumulation throughout the first 48 h of remedy (Fig. 1a, b, Supplementary Fig. 2A, Supplementary Motion pictures 1–4), which was invariably related to p27Kip1 overexpression and Retinoblastoma (Rb) protein dephosphorylation (Fig. 1c, Supplementary Fig. 2B). This sample was additionally noticed in different NSCLC fashions of oncogenic habit similar to KRASG12C, ALKEML4, or BRAFV600E, handled with their respective focused therapies (i.e., sotorasib, lorlatinib and dabrafenib) (Supplementary Fig. 3A–C). Whereas most cells remained in G1 and progressively died leading to a bulk inhabitants lower, a subset of cells, known as “early escapers”, quickly progressed by S/G2 (Fig. 1a–b, Supplementary Fig. 2A, Supplementary Fig. 3A–C, Supplementary Motion pictures 1–4), an remark in line with a uncommon, drug-tolerant biking subpopulation as not too long ago described35.
a Proportion of complete (blue), S/G2 (inexperienced) or G1 (pink) populations of HCC4006 subclonal cells throughout osimertinib remedy (1 µM). Knowledge are imply ± SEM. Consultant knowledge from n = 4 impartial organic experiments. b Consultant fluorescence pictures of S/G2 (inexperienced) or G1 (pink) HCC4006 subclonal cells throughout osimertinib remedy (1 µM) from n = 4 impartial organic experiments. Scale bar: 50 µm. c Western Blot evaluation of EGFR, retinoblastoma (Rb) and p27Kip1 signaling pathway in untreated (CT) and osimertinib-treated S/G2 or G1 HCC4006 subclonal cells. Consultant blots from n = 2 impartial organic experiments. d UMAP plot of the totally different clusters from Seurat evaluation of untreated (CT) and osimertinib-treated HCC4006 subclonal cells obtained after scRNAseq. The relative proportion of every inhabitants was G1 = 35.5% and S/G2 = 30.3% for untreated cells and G1 = 70.4% and S/G2 = 0.6% after 20 days of osimertinib remedy 1 µM. Between 2000 and 3000 cells have been recovered for every situation for library preparation. e Violin plots representing the distribution of indicated signature scores within the totally different clusters. f Dot plot exhibiting the expression degree and proportion of expressing cells of genes particular for the mucous/serous, alveolar and mesenchymal phenotypes within the totally different clusters. g, Western Blot evaluation of proteins associated to the alveolar (AGER) and mesenchymal (N-Cadherin) phenotypes in untreated, osimertinib-treated for five days and osimertinib-resistant proliferative HCC4006 subclones (RPC: Resistant Proliferative cells). Consultant blots from n = 4 impartial organic experiments. h Distribution of the imply normalized (z-score) expression of alveolar (blue), mesenchymal (pink) and E2F targets (inexperienced) signatures, based mostly on latent time. Osi-G1 cluster 4 is proven in orange and cluster 5 in blue. Knowledge are imply ± SEM. i Proliferation of G1 (pink) and S/G2 (inexperienced) HCC4006 subclonal cells sorted by FACS after 5 days of osimertinib remedy (1 µM) and re-plated within the presence of the drug. Knowledge are imply ± SEM. Consultant knowledge from n = 3 impartial organic experiments. j Imply normalized (z-score) expression per cluster of genes concerned in KRAS signaling, interferon, Rho GTPase cycle, actin cytoskeleton, NRF2 and glutathione metabolism pathways. Supply knowledge are offered as a Supply knowledge file.
To determine the molecular mechanisms underlying evolution from G1-arrest to early escape, we carried out scRNAseq on ~3000 G1 (pink) and S/G2 (inexperienced) cells sorted from each untreated and osimertinib-treated HCC4006 cells, which enabled the enrichment of the uncommon inhabitants of early escapers (Supplementary Fig. 4A, B). The seurat evaluation recognized totally different clusters, which have been largely associated to the place of cells throughout the cell cycle (Fig. 1d, Supplementary Fig. 4C). In keeping with earlier experiences19,22, myogenesis and epithelial-to-mesenchymal (EMT) signatures have been strongly upregulated in each G1- and S/G2-treated cells (Supplementary Fig. 4D), whereas cell cycle-related gene signatures similar to E2F_targets or G2M_checkpoint have been profoundly downregulated in G1 and absolutely restored in early escapers (Fig. 1e, Supp Fig. 4C, D). We noticed a deep lineage reprogramming in the course of the adaptive response, which concerned a sturdy repression of mucous/serous-related genes (e.g., PIGR, BPIFB1/2, SCGB3A1, or MUC5B) and a progressive acquisition of mesenchymal options in early escapers, in line with an EMT course of (Fig. 1e, f, Supplementary Fig. 4E–G). Curiously, certainly one of two osimertinib-treated G1 clusters (osi-G1#4) was extremely enriched in alveolar sort 1 (AT1)-gene signature45 (e.g., AQP4, CYP4B1, CLIC5, AGER or TNNC1), whereas the second G1 cluster (osi-G1#5) was enriched in mesenchymal-related genes (e.g., SERPINE1, ADAM12, SPOCK1, or MATN2) (Fig. 1e, f, Supplementary Fig. 4E–G). AT1-specific markers have been upregulated early throughout remedy and have been restricted to the non-cycling drug-tolerant inhabitants, whereas mesenchymal options have been particularly elevated in resistant proliferative cells, suggesting that tumor cells could have undergone an alveolar-like differentiation course of previous to the acquisition of a mesenchymal phenotype (Fig. 1g). The AT1-specific marker AGER was additionally transiently upregulated throughout drug-tolerance in different fashions of oncogenic habit similar to KRASG12C, BRAFV600E or ALKEML4 handled with their corresponding TT, and was misplaced in resistant proliferative cells (Supplementary Fig. 4H). Pseudotime evaluation revealed a good connection between AT1 (osi-G1#4) and mesenchymal (osi-G1#5) clusters, which means that mesenchymal-like cells may have advanced from the alveolar-like inhabitants (Fig. 1h, Supplementary Fig. 5A–C). Early escapers sorted after solely 5 days of osimertinib remedy instantly reproliferated within the presence of the drug, confirming that these cells had already acquired a resistance mechanism, whereas G1-sorted cells displayed a latency of two-to-three weeks earlier than creating resistant proliferative cells, exhibiting that this inhabitants could represent a reservoir from which resistant cells may emerge (Fig. 1i).
We subsequent aimed to find out the molecular pathways concerned within the acquisition of the totally different phenotypes. We correlated the imply expression of every gene with the expression of essentially the most consultant gene for every phenotype (e.g., AGER for the AT1 phenotype, SERPINE1 for the mesenchymal phenotype, and BPIFB1 for the mucous/serous phenotype; Pearson correlation coefficient > 0.9) (Supplementary Fig. 6A, D, G, Supplementary Knowledge 1). We decided that the mucous/serous phenotype was largely related to KRAS signaling-related genes (Fig. 1j, Supplementary Fig. 6A–C, Supplementary Knowledge 1), the alveolar phenotype was extremely enriched in interferon-related genes (Fig. 1j, Supplementary Fig. 6D–F, Supplementary Knowledge 1), and the mesenchymal-associated phenotype was strongly related to EMT, RHO_GTPASE_CYCLE and ACTIN_CYTOSKELETON signatures, in addition to some NRF2 (NFE2L2) and glutathione metabolism-related genes (Fig. 1j, Supplementary Fig. 6G–Ok, Supplementary Knowledge 1).
Altogether, our knowledge present that focused therapies invariably induce a speedy cell cycle arrest characterised by the activation of the p27Kip1/pRb pathway, adopted by the emergence of a uncommon inhabitants of proliferative early escaper cells. The drug-tolerant state displayed two distinct though tightly linked populations composed by an alveolar-like subpopulation enriched in interferon-related genes that have been restricted to the non-proliferative state, and a mesenchymal subpopulation that was largely related to early escapers and was characterised by an elevated expression of genes related to Rho-GTPase exercise and actin cytoskeleton reworking. The sequential emergence of the 2 phenotypes, first alveolar after which mesenchymal, mixed with pseudotime evaluation, counsel that mesenchymal cells could have advanced from the alveolar-like inhabitants. Nonetheless, we can not exclude that tumor cells may have undergone distinct paths of drug adaptation towards each phenotypes in the course of the adaptive response.
Drug-tolerant cells transcriptomic signature reveals similarities with regular alveolar cells
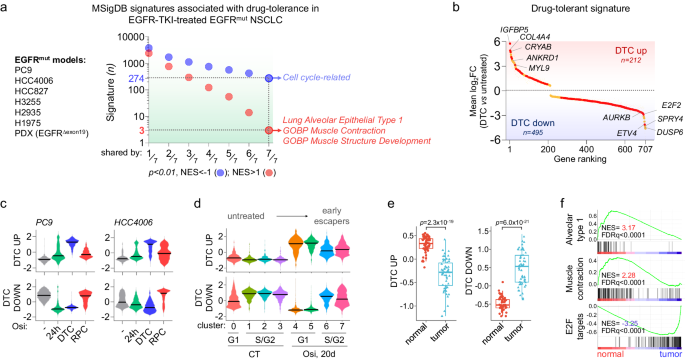
Since these findings could possibly be particular to the HCC4006 mannequin, we carried out bulk RNAseq experiments in drug-tolerant cells generated in three different EGFR-mutated cell traces handled with both erlotinib or osimertinib (PC9, HCC827, H3255), and we in contrast their transcriptomic profiles to different not too long ago revealed transcriptomes of osimertinib-induced drug-tolerance, which included two different cell traces (HCC2935 and H1975)46 and one EGFR-mutant NSCLC PDX mannequin23 (Fig. 2a, Supplementary Desk 1). Amongst 27,864 Human Molecular Signatures Database (MSigDB) gene units examined, solely 3 signatures have been generally upregulated in all of the fashions (NOM p-value < 0.05), specifically TRAVAGLINI_LUNG_ALVEOLAR_EPITHELIAL_TYPE_1_CELL, and two muscle-related gene signatures (GOBP_MUSCLE_CONTRACTION and GOBP_MUSCLE_STRUCTURE_DEVELOPMENT), with some overlapping genes between these signatures similar to MYL9, CRYAB, TAGLN, ATF3, or TNNC1 (Fig. 2a, Supplementary Fig. 7A–C, Supplementary Knowledge 2). However, 274 signatures have been generally downregulated and have been largely related to the cell cycle (Fig. 2a, Supplementary Fig. 7A, Supplementary Knowledge 2). Utilizing these transcriptomic knowledge, we constructed a brand new signature of drug tolerance composed of 212 genes generally upregulated (i.e., p < 0.01, log2FC > 0.5 in no less than 6 out of seven fashions; DTC_UP) and 495 genes generally downregulated (i.e., p < 0.01, log2FC < −0.5 in no less than 6 out of seven fashions; DTC_DOWN) (Fig. 2b, Supplementary Knowledge 3). In step with earlier evaluation, DTC_UP signature was enriched in genes associated to alveolar sort 1, EMT, muscle contraction, extracellular matrix (ECM), insulin-like progress components (IGFBPs) and RHO-GTPase_cycle (Supplementary Fig. 8A). This signature was extremely particular of the drug-tolerant state and was virtually fully reversed in absolutely resistant proliferative cells (RPC) (Fig. 2c). Constantly, DTC_UP signature was upregulated in each osi-G1#4 and osi-G1#5 clusters and was partially reversed in early escapers (Fig. second). Curiously, each DTC_UP and DTC_DOWN signatures have been strongly related to EGFR-TKI-induced drug-tolerance in vivo23 but in addition in different NSCLC fashions of oncogenic habit similar to KRASG12C 47, ALKEML4 48, or BRAFV600E 49 handled with their respective focused therapies, suggesting a standard path of drug adaptation (Supplementary Fig. 1B). DTC_UP signature was additionally related to wholesome lungs in comparison with lung adenocarcinomas, with 136/212 (64.2%) genes considerably upregulated in each DTC and regular lungs (Fig. 2e, Supplementary Fig. 9A), whereas 395 of the 495 (79.8%) genes of the DTC_DOWN signature the place considerably downregulated in regular tissue versus lung tumors (Fig. 2e, Supplementary Fig. 9B), strongly reinforcing the proof that DTC shared phenotypic resemblance with regular epithelial lung cells. However, some genes have been particularly upregulated in DTC (vs untreated) however not in wholesome lungs (vs tumors) similar to IGFBP5, SPARC, RHOBTB3, SOX4, L1CAM or TRIO, amongst others (Supplementary Fig. 9A). Conversely, some genes have been particularly downregulated in response to EGFR-TKI however not in wholesome lungs (vs tumors), similar to DUSP6, ETV5, SPRY4, PPARG, SCD of LDLR, amongst others (Supplementary Fig. 9B), highlighting variations between drug-tolerant cells and wholesome cells. As in DTC, genes overexpressed in wholesome tissues in contrast with lung adenocarcinomas correlated with AT1 and muscle contraction-related signatures (Fig. 2f, Supplementary Fig. 10A, B), and displayed enrichment for cytoskeletal-related genes (Supplementary Fig. 10A–C, Supplementary Desk 2).
a GSEA evaluation exhibiting downregulated (blue; nominal p-value < 0.01, NES < −1) and upregulated (pink; nominal p-value < 0.01, NES > 1) gene signatures from the Human Molecular Signatures Database (MSigDB), organized from the least continuously shared (1/7) to essentially the most continuously shared (7/7) signatures throughout the totally different fashions of EGFR-TKI-induced drug-tolerant cells. Nominal p-value and NES have been calculated utilizing GSEA software program. b Drug-tolerant-associated genes ranked based mostly on the imply log2 fold change expression (DTC vs untreated, p < 0.01) in 7/7 (pink) or 6/7 (orange) fashions described in a. Knowledge are imply ± SEM. p-value was obtained by DESeq2 evaluation. c and d Distribution of imply expression ranges (z-scores) of genes from the DTC_UP and DTC_DOWN signatures in untreated cells or at indicated levels of remedy in PC9 and HCC4006 subclones (c) and within the totally different clusters recognized by scRNAseq (d). e Field plots of signature scores of DTC_UP and DTC_DOWN in lung adenocarcinomas (tumor, n = 58) in comparison with adjoining regular lung tissue (regular, n = 58) from TCGA-LUAD database. The field plots show twenty fifth (decrease sure), fiftieth (middle, median), and seventy fifth (higher sure) percentiles, with whiskers generated with the Tukey technique; all factors are proven. p-value was calculated utilizing a two-tailed paired t-test. f GSEA evaluation of alveolar sort 1, muscle-contraction and E2F targets signatures in regular lungs in comparison with lung adenocarcinomas utilizing the TCGA-LUAD database. DTC: drug-tolerant cell; RPC: resistant proliferative cell. Supply knowledge are offered as a Supply knowledge file.
General, we outlined a transcriptomic signature of DTC, which revealed similarities with regular alveolar cells primarily characterised by elevated contractility and cytoskeletal reworking.
Farnesyltransferase inhibition prevents the emergence of resistance to focused therapies in NSCLC
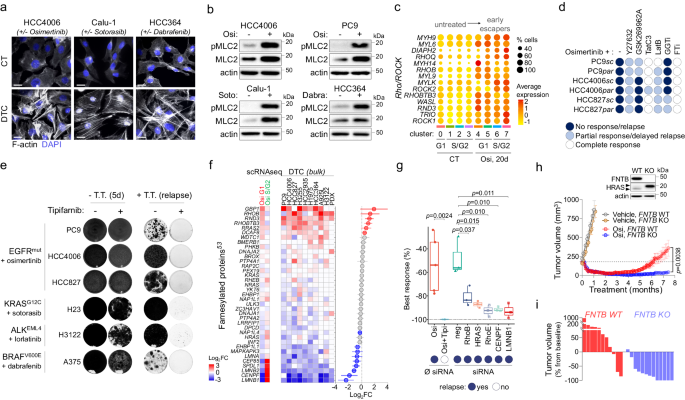
We subsequent aimed to evaluate the origin and the organic penalties of elevated contractile gene signatures. We noticed a extremely reorganized actin cytoskeleton in all of the EGFR-TKI-induced DTC evidenced by the presence of actin stress fibers and/or lamellipodia, which grew to become seen after 3 to five days of remedy (Fig. 3a, Supplementary Fig. 11A). Stress fibers have been additionally noticed in different oncogenic settings similar to KRASG12C or BRAFV600-mutant NSCLC cells handled respectively with sotorasib and dabrafenib (Fig. 3a), suggesting that actin cytoskeleton reworking was a standard mobile response to focused remedy remedy. For the reason that presence of F-actin typically correlates with Rho/ROCK pathway activation50,51, and given the affiliation of DTC with the RHO_GTPASE_CYCLE signature (Fig. 1j, Supplementary Fig. 8A), we hypothesized that this pathway may preferentially mediate the phenotypic plasticity noticed in the course of the adaptive response to focused remedy. In step with this speculation, we noticed a rise in each complete and phosphorylated myosin gentle chain 2 (pMLC2) (Fig. 3b), in addition to elevated expression of a number of Rho/ROCK-related genes in each G1 and S/G2 osimertinib-treated cells (Fig. 3c, Supplementary Fig. 11B, C). It’s noteworthy that overexpression of MYL9 (MLC2-coding gene) was noticed in each the alveolar-like and mesenchymal subpopulations (Fig. 3c, Supplementary Fig. 11D), reinforcing the phenotypic hyperlink between each subpopulations.
a Filamentous actin (F-actin) staining of indicated NSCLC cell line fashions in management or drug-tolerant cells (DTC) handled with their corresponding focused remedy at 1 µM. Scale bar: 20 µm. Consultant of n = 3 impartial organic experiments. b Western blot evaluation of complete and phosphorylated MLC2 within the indicated NSCLC cell line fashions in management cells or DTC handled with their corresponding focused remedy at 1 µM. Consultant blots from n = 3 impartial organic experiments. c Bubble plot exhibiting the expression and distribution of genes concerned within the Rho/ROCK pathway within the totally different clusters. d Mobile response of PC9, HCC4006, HCC827 parental and subclonal cells handled with 1 µM osimertinib alone or together with ROCK1/2 inhibitors (Y276324, 10 µM; GSK269962A4, 5 µM), RHOA/B/C inhibitor (TatC3, 5 µg/ml), actin polymerization inhibitor (LatB, Latruncunlin B, 0.3 µM), geranylgeranyltransferase inhibitor (GGTi, 1 µM) or farnesyltransferase inhibitor (FTi; 1 µM). Knowledge are consultant of n = 3 impartial organic experiments. e Crystal violet staining of indicated mobile fashions untreated or handled till relapse with corresponding focused remedy (T.T.) alone (1 µM) or together with Tipifarnib (1 µM). Consultant pictures from n = 3 impartial organic experiments. f Differential expression of genes (p < 0.01) coding for farnesylated proteins53 in osi-G1 vs CT-G1 (G1) and osi-S/G2 vs osi-G1 (S/G2) in HCC4006 subclonal cells (left) and in indicated NSCLC fashions of drug-tolerance (Proper); p-value for every particular person mannequin was obtained by DESeq2 evaluation. Proper: imply (Log2FC [DTC vs untreated], all fashions) ±SEM; pink dots: upregulated with p < 0.01, blue dots: downregulated with p < 0.01, grey: non-significant; p-value for imply Log2FC was calculated utilizing two-tailed unpaired t-test. g Field plots exhibiting the proportion of greatest response after remedy with osimertinib alone (1 µM) or together with Tipifarnib (1 µM), or after transfection with indicated siRNA (10 nM). The field plots show twenty fifth (decrease sure), fiftieth (middle, median), and seventy fifth (higher sure) percentiles, with whiskers generated with the Tukey technique; Knowledge signify n = 5 (osi, osi+tipi) or n = 3 (siRNA) impartial organic experiments. p-value was calculated utilizing a two-tailed unpaired t-test. h Prime: Western blot evaluation of FNTB protein expression and HRAS prenylation standing within the FNTB WT and KO cells. The higher and decrease arrows present unfarnesylated and farnesylated proteins respectively. Backside: Imply tumor quantity of PC9 FNTB-WT and FNTB-KO xenografts. Mice have been handled 5 days/week with automobile or Osimertinib (Osi, 5 mg/kg, q.d). Automobile WT, n = 3; Automobile KO, n = 3; osi WT, n = 12; osi KO, n = 14. Knowledge are imply ± SEM; the p-value was calculated utilizing two-tailed unpaired t-test. i Proportion of tumor quantity from baseline after 7 months of osimertinib remedy in mice bearing FNTB-WT and FNTB-KO PC9 xenografts. Supply knowledge are offered as a Supply knowledge file.
Primarily based on these observations, we aimed to find out whether or not pharmacological inhibition of this pathway may stop the emergence of resistance to EGFR-TKI. We examined a panel of inhibitors which included ROCK1/2 inhibitors similar to Y27632 and GSK269962, the RhoA/B/C inhibitor C3-exoenzyme (tat-C3). For the reason that exercise of a number of RhoGTPases upregulated throughout drug tolerance (Fig. 3c, Supplementary Fig. 11B and C) depends upon their prenylation standing52, we additionally included a farnesyltransferase inhibitor (FTI, tipifarnib) and a geranylgeranyltransferase inhibitor (GGTi, GGTi-298). Inhibitors alone didn’t have an effect on cell proliferation, aside from the HCC4006 cell line by which tipifarnib displayed a cytostatic exercise (Supplementary Fig. 12A). Curiously, all these inhibitors interfered with the traditional course of the adaptive response to EGFR-TKI, though they confirmed totally different exercise profiles. As an example, ROCK inhibitors impaired osimertinib-induced MLC2 activation and stress fiber formation (Supplementary Fig. 12B-C) however did not remove DTC and fairly promoted a long-term stabilization of the G1 inhabitants (Fig. 3d, Supplementary Fig. 12A and D), along with an elevated expression of the AT1-specific marker AGER (Supplementary Fig. 12B). This means that ROCK activation could also be required for mesenchymal transition however is dispensable for AT1 non-cycling cell survival. RhoA/B/C inhibitor tat-C3 additionally decreased stress fiber formation (Supplementary Fig. 12C) and strongly elevated EGFR-TKI effectivity, nevertheless some resistant clones may nonetheless be noticed relying on the cell sort (Fig. 3d, Supplementary Fig. 12A). Curiously, amongst all of the inhibitors examined, the FTI tipifarnib was the one drug that fully prevented the event of resistances to each osimertinib and erlotinib in all of the cell traces examined (Fig. 3d and e, Supplementary Fig. 12A). Remarkably, tipifarnib additionally prevented relapse to focused therapies in different fashions of oncogenic habit similar to ALKEML4 and KRASG12 NSCLC or BRAFV600E-mutant melanoma cell traces handled with lorlatinib, sotorasib and dabrafenib, respectively (Fig. 3e, Supplementary Fig. 13A). Different FTIs similar to CP-609754 or lonafarnib equally prevented relapse to osimertinib, confirming that the impact was class-wide and never restricted to a single drug (Supplementary Fig. 13B). Tipifarnib additionally induced cell demise when administered after early relapse (Supplementary Fig. 13C), whereas absolutely resistant proliferative clones displayed totally different levels of sensitivity to tipifarnib with some exhibiting an virtually full response and a few others an entire lack of sensitivity (Supplementary Fig. 13D). These outcomes counsel that tipifarnib preferentially interferes with the earliest levels of drug adaptation, though it could retain some exercise in absolutely resistant proliferative cells relying on the clone.
We subsequent aimed to find out which farnesylated protein(s) may mediate each cell survival and phenotypic reorganization of tumor cells in response to TT. We first assessed mRNA expression ranges of the genes coding for farnesylated proteins (40 in response to a current examine53). Our scRNAseq knowledge confirmed that whereas farnesylated GTPases similar to RND3 (RHOE), RHOB, or RHOBTB3 have been overexpressed by osimertinib throughout G1 arrest, the expression of a number of cell-division-related genes whose protein merchandise are farnesylation-dependent, similar to lamins (LMNB1, LMNB2, LMNA), centromere protein F (CENPF) or spindle equipment coiled-coil protein 1 (SPDL1) was extremely repressed throughout G1 however absolutely restored in S/G2 (Fig. 3f, left). This sample was extremely conserved amongst a number of fashions of DTC, suggesting that this steadiness of expression amongst farnesylated proteins could possibly be a trademark of drug tolerance (Fig. 3f, proper). Nonetheless, though siRNA-mediated inhibition of particular person farnesylated proteins similar to RHOB, RHOE, LMNB1, CENPF or HRAS54 considerably elevated sensitivity to TT, none may absolutely recapitulate tipifarnib means to current relapse (Fig. 3g), which may counsel a multi-target mode of motion for FTIs. To exclude potential off-target results of tipifarnib, we knocked out FNTB, which codes for the beta subunit of farnesyltransferase, in PC9 cells (Supplementary Fig. 14A). FNTB depletion strongly prevented protein farnesylation as decided by a attribute shift of the HRAS protein (Supplementary Fig. 14A–C) just like the shift noticed in response to tipifarnib (Supp Fig. 14D). Strikingly, in vivo osimertinib remedy of mice bearing xenograft tumors from FNTB-KO PC9 cells led to a sturdy response for the complete 7.5-month remedy interval (12/13, 92.3%), whereas most FNTB-WT PC9 tumors (8/11, 72.7%) had relapsed inside this timeframe (Fig. 3h, i). Importantly, FNTB depletion didn’t have an effect on tumor progress in vehicle-treated mice, indicating that the impact was particular to osimertinib remedy (Fig. 3h). Comparable outcomes have been obtained in vitro with each FNTB-KO and FNTB-KD PC9 cells, suggesting that partial inhibition of the farnesyltransferase was enough to change the emergence of osimertinib-resistant cells (Supplementary Fig. 14E).
Lastly, we aimed to check the flexibility of tipifarnib to forestall TT-induced stress-fiber formation, and we noticed divergent results amongst the fashions (Supplementary Fig. 15A). In Calu-1 KRASG12C-mutant cell line, tipifarnib strongly prevented sotorasib-induced MLC2 phosphorylation and stress-fiber formation (Supplementary Fig. 15A–C), and strongly prevented RhoE farnesylation as decided by a attribute shift (Supplementary Fig. 15B). On this mannequin, siRNA-mediated downregulation of RHOE strongly prevented pMLC2 and stress-fiber formation (Supplementary Fig. 15D–E), and likewise elevated sensitivity to sotorasib (Supplementary Fig. 15F). Nonetheless, though RHOE downregulation may sensitize tumor cells in different oncogenic settings similar to osimertinib-treated HCC4006 cells (Fig. 3g), its means to modulate cytoskeleton reorganization was both moderated or not noticed in different fashions, suggesting that different proteins could promote stress-fiber formation in response to the mobile context.
General, we decided that drug tolerance is invariably related to Rho/ROCK-mediated stress-fiber formation, though this phenotypic attribute was not essential for DTC survival as highlighted by co-treatment with ROCK inhibitors, however is perhaps required for cell re-proliferation. We additionally recognized tipifarnib as a extremely environment friendly drug in stopping relapse to a broad vary of TT, though its efficacy doesn’t appear to rely on its means to modulate ROCK exercise, however extra seemingly includes the inhibition of a number of farnesylated proteins that show a extremely conserved sample of regulation among the many drug-tolerant fashions.
Osimertinib-tipifarnib (OT) co-treatment induces mitotic defects and ISR-mediated apoptotic pathway
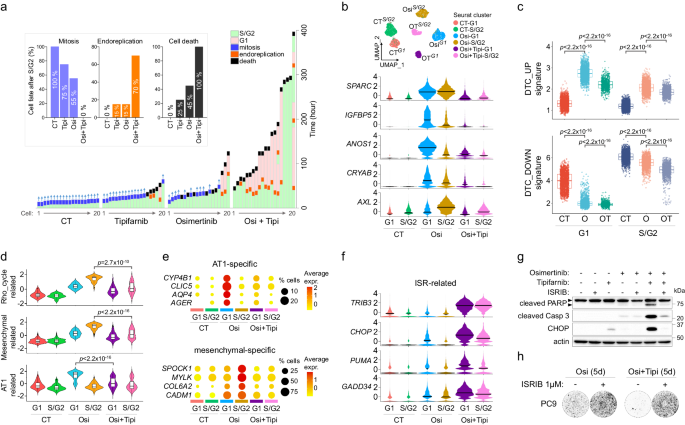
We first aimed to decipher the physiological consequence of OT remedy on DTC. Time-lapse imaging revealed that though tipifarnib didn’t stop the emergence of osimertinib-derived early escapers, escaping cells did not bear mitosis and in the end died (Fig. 4a), suggesting that OT remedy could intervene with cell division, which was in line with an inhibition of cell-cycle associated proteins similar to lamins, CENPF or RhoGTPases, amongst others (Fig. 3f). We additionally noticed {that a} excessive proportion of OT-treated cells that progressed to S/G2 returned to G1 with out dividing, in line with a technique of mitotic slippage or endoreplication55,56, which was additionally observable at decrease frequency in osimertinib-treated cells (Fig. 4a, Supplementary Fig. 16, Supplementary Film 5).
a Cell destiny after development to S/G2 of HCC4006 subclones handled or not with osimertinib (1 μM), tipifarnib (1 μM) or the mixture (Osi+Tipi). Backside: The information represents the monitoring of cells (n = 20/situation) since their entry to S/G2 section, with the time spent for every section/occasion of the cell cycle. Prime: The bar plot recapitulates the proportion of cells that skilled mitosis and endoreplication after S/G2 section, and the proportion of cells that died earlier than dividing. b Prime: UMAP plot of the totally different clusters of untreated, osimertinib-treated and osimertinib+tipifarnib-treated G1 and S/G2 HCC4006 subclones obtained by scRNAseq. Backside: Distribution of normalized expression ranges (z-score) of DTC-related genes regulated by tipifarnib-osimertinib co-treatment. c Signature scores of DTC_UP and DTC_DOWN in untreated (CT), osimertinib (O) and osimertinib + tipifarnib (OT) handled G1 and S/G2 cells. The field plots show twenty fifth (decrease sure), fiftieth (middle, median), and seventy fifth (higher sure) percentiles, with whiskers generated with the Tukey technique; all factors are proven. p-value was calculated utilizing a two-sided Wilcoxon take a look at. d Signature scores of RHO_GTPase_cycle-related, mesenchymal-related and alveolar sort 1 (AT1)-related gene signatures in untreated (CT), osimertinib (Osi) and osimertinib + tipifarnib (Osi+Tipi) handled G1 and S/G2 cells. The field plots inside violin plots are just like c. p-value was calculated utilizing two-sided Wilcoxon take a look at. For c and d: CT-G1, n = 1678; O-G1, n = 2097, OT-G1, n = 897; CT-SG2, n = 2654; O-SG2, n = 1963; OT-SG2, n = 1266. e Dot plot exhibiting the expression degree and proportion of cells expressing genes particular for the alveolar and mesenchymal phenotypes within the totally different clusters. f Distribution of normalized expression ranges (z-score) of genes associated to built-in stress response (ISR) pathway. g Western blot evaluation of proteins associated to apoptosis (PARP and caspase-3) and ISR (CHOP) of HCC4006 subclones handled with osimertinib (1 μM), tipifarnib (1 μM) and ISR inhibitor (ISRIB, 1 µM) alone or together. Consultant blots from n = 3 impartial organic experiments. The higher arrow reveals the overall and the decrease arrow reveals cleaved PARP. h Crystal violet staining of PC9 cells pre-treated or not with ISRIB (1 µM, 24 h) and handled for five days with 1 µM osimertinib alone or together with 1 µM tipifarnib. Consultant pictures from n = 3 impartial organic experiments. Supply knowledge are offered as a Supply knowledge file.
To decipher the molecular mechanisms underlying the flexibility of tipifarnib to forestall relapse to osimertinib, we carried out scRNAseq on OT-treated G1 and S/G2 sorted cells. OT-treated cells displayed a definite transcriptomic profile in comparison with osimertinib-treated cells (Fig. 4b, Supplementary Fig. 17A). Strikingly, co-treatment with tipifarnib strongly prevented osimertinib-induced overexpression of most genes related to drug-tolerance, similar to SPARC, IGFBP5, ANOS1, CRYAB or AXL (Fig. 4b), leading to a big downregulation of DTC_UP and DTC_DOWN signatures in each G1 and S/G2-treated cells (Fig. 4c). Certainly, OT remedy strongly interfered with RHO_GTPASE_CYCLE, AT1 and mesenchymal-related signatures (Fig. 4d, e, Supplementary Fig. 17B). Constantly, interferon and EMT signatures have been essentially the most considerably downregulated pathways in G1 and S/G2 OT-treated cells, respectively (Supplementary Fig. 17C–E). Lastly, we sought to find out the molecular mechanisms answerable for cell demise below OT co-treatment. We noticed a powerful enrichment in unfolded protein response (UPR)/built-in stress response (ISR)-related gene signatures in each G1 and S/G2 OT-treated cells (Supplementary Fig. 18A, B), which was concordant with a powerful overexpression of ATF4-regulated genes similar to DDIT3/CHOP, TRIB3, CHAC1, PSAT1, FAM129A/NIBAN1, PPP1R15A/GADD34 or the pro-apoptotic BBC3/PUMA, amongst others (Fig. 4f). Upregulation of ATF4 and CHOP by OT remedy was confirmed on the protein degree and correlated with PARP and Caspase 3 cleavage (Supplementary Fig. 18C). Most significantly, pharmacological inhibition of ISR utilizing ISRIB strongly prevented cell demise (Fig. 4g, h), confirming that activation of this pathway was answerable for OT-induced cell demise. Curiously, inhibition of ISR additionally prevented sotorasib+tipifarnib-induced cell demise in KRASG12C fashions (H23 and Calu-1) and lorlatinib+tipifarnib within the ALKEML4 mannequin H3122, suggesting a standard mode of motion of tipifarnib in different oncogenic settings (Supplementary Fig. 18D).
General, we noticed that co-treatment with tipifarnib induces deadly mitotic defects when mixed with osimertinib, and broadly interferes with the drug-tolerant state. OT remedy resulted within the activation of the ATF4/CHOP stress response pathway, which resulted within the apoptosis of co-treated cells.
Tipifarnib prevents relapse to focused therapies in vivo
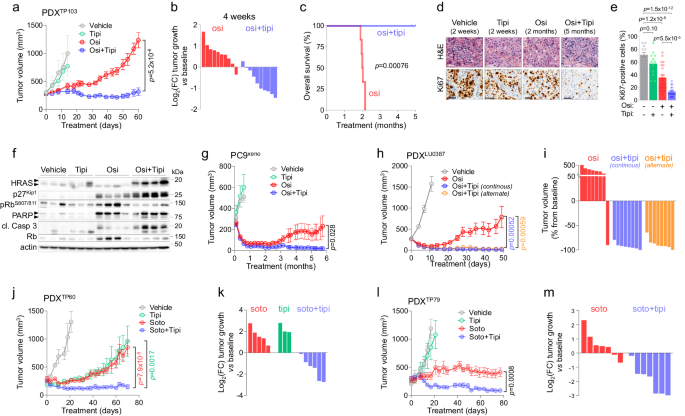
Lastly, we aimed to translate our findings to pre-clinical in vivo fashions by testing the mixture of tipifarnib with totally different focused therapies. We first carried out osimertinib and OT therapies in two totally different EGFR-mutant NSCLC PDX fashions harboring respectively an EGFRL858R/T790M double mutation (mannequin TP10357) and an exon 20 insertion (mannequin LU0387)58, in addition to in a PC9 xenograft mannequin. For all three fashions, OT co-treatment confirmed higher anti-tumor efficacy than osimertinib alone, and most significantly, the addition of tipifarnib strongly and durably prevented the emergence of osimertinib resistance, with no proof of toxicity as proven by steady physique weight and good common side of mice (Fig. 5, Supplementary Fig. 19B, F, G and 20D, F, G). Within the TP103 mannequin, osimertinib induced a average response and relapse was noticed for all (10/10) mice inside 2 months, whereas the addition of tipifarnib induced considerably larger tumor regression, with an virtually steady illness lasting the complete dosing interval of 5 months (Fig. 5a–c, Supplementary Fig. 19A, D, E, H). In step with in vitro knowledge, OT-treated tumors confirmed a strongly lowered proliferative state even after 5 months of remedy as highlighted by Ki67 staining (Fig. 5d, e). This correlated with elevated p27Kip1 ranges and lowered phospho-Rb, in addition to elevated PARP and Caspase 3 cleavage, suggesting that non-cycling OT-treated cells have been present process apoptosis (Fig. 5f). Tipifarnib-mediated farnesyltransferase inhibition was highlighted by a attribute shift of the HRAS protein towards a non-farnesylated state (Fig. 5f). Comparable outcomes have been obtained within the PC9 xenograft mannequin, the place OT remedy induced a powerful, steady and apparently secure response for as much as 6 months, with virtually full tumor regression in all animals, whereas osimertinib induced a much less potent impact and relapse was noticed in 4/10 tumors (Fig. 5g, Supplementary Fig. 20A–D). For the LU0387 mannequin, tipifarnib was administered both constantly or intermittently (1 week on, 1 week off), to higher replicate the remedy routine typically utilized in scientific follow for this molecule54. Regardless of a powerful preliminary anti-tumor impact of osimertinib, 7/8 (87.5%) tumors relapsed inside 20 days of remedy, whereas each steady and intermittent OT therapies induced stronger tumor regression and a steady response in 16/16 tumors (100%) for the complete 50-days remedy interval (Fig. 5h and that i, Supplementary Fig. 20E). Notably, tipifarnib alone didn’t show anti-tumor exercise when used alone within the TP103 and PC9 xenograft fashions (and was not assessed in LU0387 PDX), highlighting an artificial lethality between osimertinib and tipifarnib within the context of the adaptive response to TT (Fig. 5a and g, Supplementary Fig. 19A, D, E, H and 20C).
a Imply tumor quantity of the TP103 NSCLC PDX mannequin (EGFRL858R/T790M) handled 5 days/week with automobile (n = 5), tipifarnib (tipi, 80 mg/kg, b.i.d., n = 5), osimertinib (osi, 5 mg/kg, q.d, n = 10) or by the mixture (osi+tipi, n = 10). Knowledge are imply ± SEM; p-value was calculated utilizing two-tailed unpaired t-test. b Log2FC tumor progress vs baseline at 4-week remedy with osimertinib or osimertinib+tipifarnib. c General survival of the mice handled with osimertinib or osimertinib+tipifarnib. The graph is the results of one cohort of mice with n = 6 mice in each arms. p-value was calculated utilizing the log-rank Mantel-Cox take a look at. d Consultant pictures of Hematoxylin and Eosin (H&E) staining and Ki67 IHC from PDX tumors collected after indicated occasions and coverings. Knowledge are consultant of n = 3 (automobile, tipifarnib), n = 6 (osimertinib) and n = 5 (osimertinib+tipifarnib) impartial tumors. Scale bar: 50 µm. e Ki67 IHC scores quantified from 4 totally different zones of every impartial tumor introduced in d. f Western Blot evaluation of particular person PDX after 2 weeks (automobile, n = 3; tipifarnib, n = 3), 2 months (osimertinib, n = 4) and 5 months (osi+tipi, n = 4) remedy. For HRAS: the higher arrow reveals unfarnesylated and the decrease arrow reveals farnesylated protein; for PARP: the higher arrow reveals complete and the decrease arrow reveals cleaved protein. g Imply tumor quantity of PC9 xenografts handled 5 days/week with automobile (n = 6), tipifarnib (Tipi, 80 mg/kg, b.i.d., n = 6), osimertinib (osi, 5 mg/kg, q.d, n = 10), or by the mixture (osi+tipi, n = 12). Knowledge are imply ± SEM; p-value was calculated utilizing two-tailed unpaired t-test. h Imply tumor quantity of the LU0387 NSCLC PDX mannequin (EGFRexon20 insertion) handled 5 days/week with automobile (n = 8), osimertinib (osi, 25 mg/kg, q.d, n = 8), or the mixture (osimertinib + tipifarnib at 60 mg/kg, constantly b.i.d, n = 8, or intermittently 1 week ON/1 week OFF, n = 8). Knowledge are imply ± SEM; p-value was calculated utilizing two-tailed unpaired t-test. p-value (osi+tipi steady vs osi) is proven in blue and (osi+tipi alternate vs osi) in orange. i Proportion of LU0387 tumor quantity vs baseline at day 49. j Imply tumor quantity of the TP60 NSCLC PDX mannequin (KRASG12C) handled with automobile (n = 3), tipifarnib (tipi, 80 mg/kg, b.i.d., n = 3), sotorasib (soto, 30 mg/kg, q.d, n = 5), or the mixture (soto+tipi, n = 6). Knowledge are imply ± SEM; p-value was calculated utilizing two-tailed unpaired t-test. p-value (soto+tipi vs soto) is proven in pink and (soto+tipi vs tipi) in inexperienced. ok Log2 fold change of the TP60 PDX tumor dimension in comparison with baseline at day 70. l Imply tumor quantity of the TP79 NSCLC PDX mannequin (KRASG12C) handled with automobile (n = 6), tipifarnib (tipi, 80 mg/kg, b.i.d., n = 4), sotorasib (soto, 30 mg/kg, q.d, n = 7), or the mixture (soto+tipi, n = 7). Knowledge are imply ± SEM; p-value was calculated utilizing two-tailed unpaired t-test. m Log2 fold change of the TP79 PDX tumor dimension in comparison with baseline at day 77. Supply knowledge are offered as a Supply knowledge file.
Primarily based on our in vitro ends in KRASG12C-mutant fashions, we aimed to judge the mixture of tipifarnib with sotorasib in two totally different KRASG12C-mutant NSCLC PDX fashions (TP60 and TP79)59. Strikingly, combinatory remedy induced a a lot stronger and sturdy anti-tumor response in each fashions in comparison with sotorasib alone (Fig. 5j–m), with a steady response throughout the complete dosing interval of 70 days for TP60 (Fig. 5j) and 77 days for TP79 (Fig. 5l). As with the mixture with osimertinib, the tipifarnib and sotorasib pairing was well-tolerated as indicated by steady physique weight and good common side of mice (Supplementary Fig. 20F, G).
Altogether, our in vivo knowledge present that tipifarnib safely and durably prevents the emergence of resistances to osimertinib in EGFR-mutant and to sotorasib in KRASG12C-mutant NSCLC, thus offering a powerful rationale to judge these combos within the clinic.