We analysed 10,478 most cancers genomes spanning 35 totally different most cancers varieties (Fig. 1b and Supplementary Tables 1 and 2). Whereas broadly reflecting the spectrum and frequencies of cancers identified within the UK inhabitants, there have been variations, with an over-representation of colorectal and kidney cancers and a paucity of prostate and pancreatic cancers (Prolonged Information Fig. 1). Moreover, for the principle most cancers varieties, the sufferers recruited to 100kGP tended to be youthful and had earlier stage tumors in comparison with sufferers within the normal UK inhabitants (Supplementary Desk 3).
Mutation charges various throughout the totally different most cancers varieties with cutaneous melanoma having the best single nucleotide variant mutation rely and meningioma the bottom (Prolonged Information Fig. 2). A complete of 945 samples, notably colorectal and uterine cancers, had been hypermutated, both as results of faulty mismatch restore (dMMR) or POLE mutation. Invasive ductal carcinoma of the breast had the best energy for driver gene detection (>90% energy for a mutation fee of at the least 2% increased than background) and enormous cell lung most cancers had the bottom energy (Fig. 2 and Supplementary Desk 4). In contrast with the latest Pan-Most cancers Evaluation of Complete Genomes evaluation12, the 100kGP cohort was higher powered to establish a driver mutation for 19 cancers, notably for breast, colorectal, esophageal and uterine most cancers, lung adenocarcinoma and bladder transitional cell carcinoma the place the pattern sizes had been greater than tenfold increased.
Spectrum of most cancers driver genes
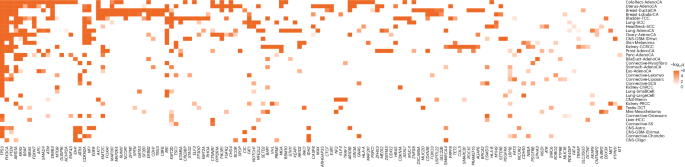
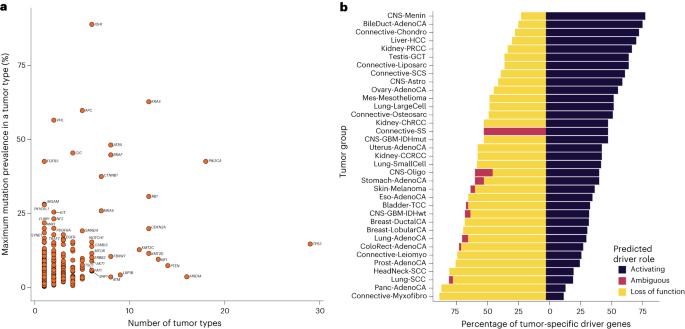
Throughout all most cancers varieties we recognized 770 distinctive tumor–driver gene pairs akin to 330 distinctive candidate most cancers driver genes (Fig. 3, Prolonged Information Fig. 3 and Supplementary Desk 5). When in comparison with the most important pan-cancer driver evaluation, in 21 of 31 most cancers varieties the place tumor histologies might be matched, we recovered 61% of all most cancers drivers reported by the Catalogue of Somatic Mutations in Most cancers (COSMIC), the Integrative OncoGenomics (IntOGen)4 and The Most cancers Genome Atlas (TCGA) Program pan-cancer evaluation reported by ref. 13 (Supplementary Desk 5). We had been capable of detect 80% of drivers reported for colorectal, breast, lung and ovarian cancers however solely <20% of drivers reported for hepatocellular and abdomen cancers, which can be a results of differing pattern dimension or intertumour heterogeneity14. The variety of recognized most cancers driver genes various between most cancers varieties, with colorectal and uterine cancers having probably the most (60 genes) and spindle cell carcinoma having the fewest (4 genes). Throughout the 35 cancers, we discovered no correlation between common mutation burden and the variety of driver genes in every most cancers (Pearson’s r = 0.19, P = 0.27). The consensus checklist additionally consists of 326 tumor–driver pairs that haven’t beforehand been reported by the Most cancers Gene Census, IntOGen or the pan-cancer evaluation of TCGA4,13 (Supplementary Desk 5) and 74 that haven’t beforehand been related to any particular tissue. Virtually all the candidate drivers recognized had been unusual, with 88% (65 of 74) having a mutation frequency <10% within the respective most cancers sort. The best numbers of latest most cancers driver genes had been discovered for uterine (n = 42), bladder (n = 40) and colorectal (n = 37) cancers. Moreover, we recognized drivers in tumor varieties which haven’t been cataloged by IntOGen4 and ref. 13. These embrace breast lobular carcinoma, meningioma and myxofibrosarcoma. Predictions of identified most cancers driver genes in new most cancers varieties embrace SPTA1, CHD4 and ASXL1 in colorectal most cancers, FOXO3, MUC16 and ZFPM1 in breast cancers and CNTNAP2, CTNND2 and TRRAP in lung adenocarcinoma. Fully new predictions embrace MAP3K21 (encoding a mixed-lineage kinase) in colorectal most cancers, USP17L22 (encoding a deubiquitinating enzyme) in breast ductal carcinoma and TPTE (encoding a tyrosine phosphatase) in lung adenocarcinoma (Supplementary Desk 5).
Eighty-five genes had been recognized as a driver in additional than two tumor varieties, with 26 genes functioning as drivers in additional than 5 tumor varieties (Fig. 4a). As anticipated, TP53 was recognized as a driver gene in probably the most tumor varieties, adopted by PIK3CA, ARID1A and PTEN, appearing as most cancers driver genes in 29, 18, 16 and 14 totally different tumor varieties, respectively. Whereas many genes operate as drivers in a number of most cancers varieties, some drivers are mutated at excessive frequencies solely in particular tumors, resembling VHL in clear cell renal cell carcinoma and FGFR3 in bladder most cancers (Fig. 4a). Throughout drivers working in a number of most cancers varieties, the clearest examples of domain-specific driver mutations had been in EGFR, the place protein tyrosine and serine/threonine kinase area mutations predominated in lung adenocarcinoma, in distinction to extracellular furin-like cysteine-rich area area mutations in IDH wild-type glioblastoma (Supplementary Desk 6 and Prolonged Information Fig. 4a). PIK3CA additionally confirmed a choice for p85-binding area mutations in uterine adenocarcinoma in comparison with different most cancers varieties, resembling breast ductal carcinoma, that are enriched for mutations within the PIK household area (Supplementary Desk 6 and Prolonged Information Fig. 4b). Hierarchical clustering of cancers primarily based on the presence of recognized driver mutations and their respective q worth demonstrated clustering of most cancers varieties by cell of origin (for instance, head and neck and lung squamous cell carcinoma) and by organ (for instance, breast ductal and lobular carcinomas; Prolonged Information Fig. 5). The ratio of predicted activating versus tumor suppressor driver genes various throughout tumor varieties with meningioma and myxofibrosarcoma possessing the best and lowest ratios, respectively (Fig. 4b and Supplementary Desk 5).
a, Distribution of driver genes throughout several types of most cancers: y axis, maximal mutational prevalence in a tumor sort; x axis, variety of tumor varieties by which the motive force gene is recognized. Genes labeled are candidate drivers in at the least six tumor varieties or have a most mutation prevalence in a tumor sort of >17%. b, Distribution of most cancers driver gene operate related to every most cancers sort: y axis, tumor group; x axis, proportion of tumor-specific driver genes.
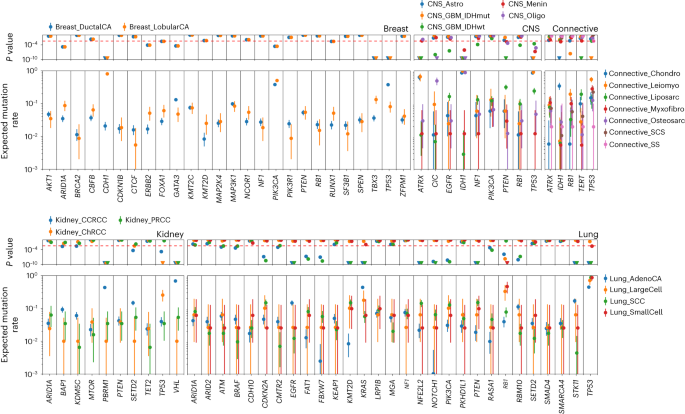
Throughout the 35 totally different tumor varieties in 9,070 distinctive samples we recognized 12,606 distinct oncogenic mutations in tumor-relevant most cancers driver genes. The median variety of oncogenic mutations in most cancers driver genes per pattern was two, throughout all tumors. The best median variety of oncogenic mutations in driver genes per pattern was seen in uterine most cancers (n = 6; Prolonged Information Fig. 6). We noticed vital variations (Pbinomial < 3.5 × 10−3) in oncogenic mutation frequency in most cancers driver genes throughout totally different tumor histologies arising from the identical organ. Examples embrace CDH1, TBX3 and TP53 in breast cancers, ATRX, CIC, IDH1, PTEN and TP53 in central nervous system tumors, IDH1 and TP53 in connective tissue tumors, PBRM1 and VHL in renal cancers and EGFR, KMT2D, KRAS, NFE2L2, PTEN, STK11 and TP53 in lung cancers (Fig. 5).
Anticipated mutation fee and 95% confidence intervals of every driver within the cohort (2,306 breast, 440 central nervous system (CNS), 1,045 kidney, 1,110 lung and 607 connective tissue tumors within the 100kGP cohort) primarily based on the variety of samples by which the motive force gene is mutated for the given tumor histology. Binomial P values are proven. The dashed pink line corresponds to a false discovery fee of 0.01.
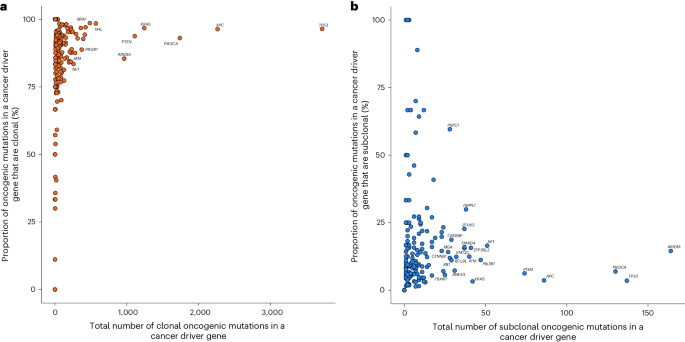
Contemplating all 330 most cancers driver genes, 217 featured at the least one clonal oncogenic mutation (214 clonal, 167 clonal early and 114 clonal late occasions (Supplementary Desk 7). APC, TP53 and PIK3CA possessed probably the most clonal oncogenic mutations (Fig. 6a and Prolonged Information Fig. 7). Of the 162 driver genes that harbored at the least one subclonal oncogenic mutation, ARID1A, TP53 and PIK3CA possessed probably the most (Fig. 6b and Prolonged Information Fig. 7). Per revealed work, a excessive proportion (55%) of all early clonal driver mutations happen in simply 4 genes (TP53, APC, KRAS and PIK3CA) whereas the equal proportion of late and subclonal oncogenic mutations was noticed in 19 totally different genes (Supplementary Desk 7)15,16,17,18. This discovering helps a mannequin by which early occasions in most cancers evolution are likely to happen in a restricted set of driver genes and a wider vary of drivers characteristic late in tumor evolution. In tumors with greater than ten oncogenic mutations, meningioma exhibited the best proportion of clonal oncogenic mutations (Prolonged Information Fig. 8a). Giant cell lung, testicular germ cell tumor and oligodendroglioma carried the best proportion of early clonal, late clonal and subclonal oncogenic mutations, respectively (Prolonged Information Fig. 8b–d).
a, Distribution of clonal oncogenic mutations in candidate most cancers driver genes throughout all most cancers varieties: y axis, proportion of all clonal oncogenic mutations of all oncogenic mutations; x axis, whole variety of clonal oncogenic mutations. Clonal oncogenic mutations embrace clonal mutations that occurred earlier than duplications involving the related chromosome (early), clonal mutations that occurred after such duplications (late), and mutations that occurred when no duplication was noticed. Genes labeled are these with >250 clonal oncogenic mutations or clonal oncogenic mutations symbolize >95% of all oncogenic mutations. b, Distribution of all subclonal oncogenic mutations in candidate most cancers driver genes throughout all most cancers varieties: y axis, proportion of all subclonal oncogenic mutations of all oncogenic mutations; x axis, whole variety of subclonal oncogenic mutations. Genes labeled are these with >50 subclonal oncogenic mutations and >5% of all oncogenic mutations as subclonal.
Sensitivity of WGS mutation detection in comparison with panels
We initially investigated the efficiency of WGS to detect clinically related mutations in comparison with typical panel-based testing by way of comparability of mutation calls with Memorial Sloan Kettering (MSK) Most cancers Middle cohorts at 43 established drivers (Supplementary Observe 1). For major tumors represented within the MSK and 100kGP cohorts, the speed of mutations referred to as for every driver gene was comparable (Supplementary Figs. 1 and 2). Thereafter, we estimated the sensitivity of mutation detection within the 100kGP cohort by extracting per-tumor protection throughout the panel of 43 driver genes (Supplementary Observe 1). Particularly, for 88% of most cancers driver genes, the anticipated sensitivity for mutation detection was >99% within the 100kGP cohort. Moreover, for 90% of most cancers driver genes, >98% of the coding sequence had adequate protection such that greater than six reads might be used for mutation detection after accounting for tumor purity (Supplementary Figs. 3–7). These findings are in settlement with revealed information on the diagnostic accuracy of 100kGP WGS in comparison with panel sequencing carried out by Genomics England (sensitivity of 99% for variant allele frequency >5% and protection >70×).
Actionability of driver gene mutations
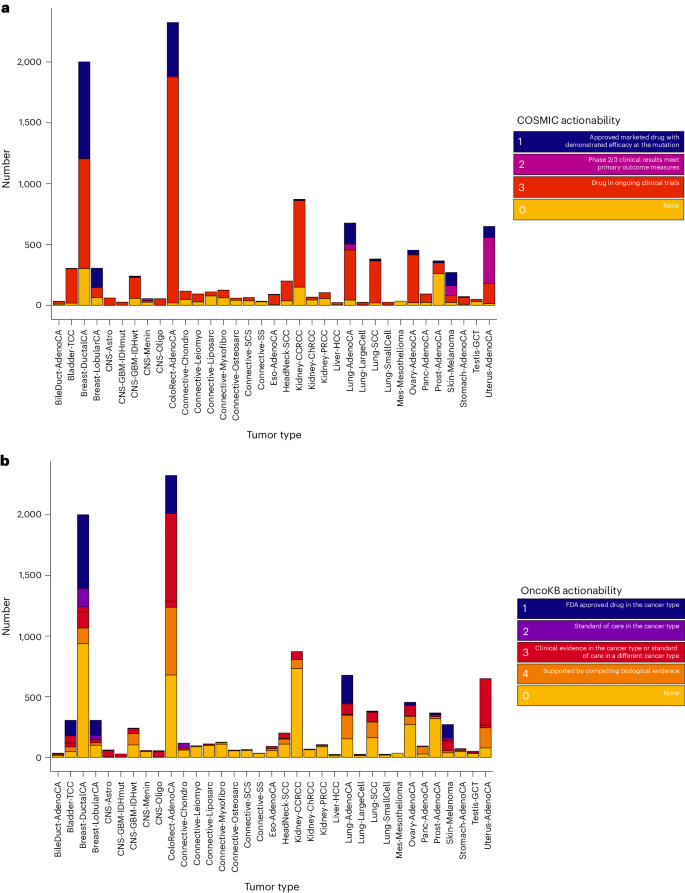
We subsequent sought to guage the panorama of clinically actionable driver alterations by way of reference to the COSMIC and Precision Oncology Data Base (OncoKB). We noticed that each the fraction of samples and proportion of alteration varieties various throughout tissue varieties. Information from COSMIC indicated that 85% of all samples (8,880 of 10,478) possessed at the least one putatively actionable alteration being focused in a scientific setting (Fig. 7a and Supplementary Desk 8), whereas 55% of samples (5,805 of 10,478) had at the least one putatively actionable or biologically related alteration from OncoKB (Fig. 7b and Supplementary Tables 9 and 10). Throughout all most cancers varieties, 15% (1,560 of 10,470) of the sufferers could be eligible for a presently accredited remedy as outlined by OncoKB. Of the actionable mutations outlined by OncoKB (n = 9,639), 5,823 had been clonal, 2,632 had been early clonal, 229 had been late clonal and 852 had been subclonal.
a, Scientific actionability ascribable to every candidate most cancers driver gene in line with COSMIC by most cancers sort. Tumors had been annotated by the best scoring gene mutation–indication pairing, with ‘None’ indicating no actionable mutations had been detected within the tumor. b, Scientific actionability ascribable to every candidate most cancers driver gene in line with OncoKB by most cancers sort. Tumors had been annotated by the best scoring gene mutation–indication pairing, with ‘None’ indicating no actionable mutations had been detected within the tumor.
The commonest putatively actionable alterations throughout all the 35 most cancers varieties had been mutations in PIK3CA, KRAS and PTEN (Supplementary Fig. 8). PIK3CA encodes the p110α protein, which is a catalytic subunit of phosphatidylinositol 3-kinase (PI3K). Particular oncogenic missense mutations in PIK3CA had been current in 50% of lobular breast cancers and 38% of ductal breast cancers and their presence is a sign for the usage of PI3Kα inhibitor alpelisib19. These mutations are current in a variety of cancers together with colorectal (20%) and uterine cancers (47%) and in these tumor varieties are topic to early scientific research with an allosteric inhibitor of PI3Kα20. We discovered excessive fractions of sufferers with pancreatic most cancers, colorectal most cancers and lung adenocarcinoma with actionable KRAS mutations (39–64% of all instances). The KRAS G12C mutation was current in 17% of lung adenocarcinoma instances and is focused by mutation-specific selective covalent inhibition with adagrasib or sotorasib21,22. PI3Kβ inhibition is of great organic curiosity in sufferers with oncogenic inactivating PTEN mutations, as PI3Kβ is assumed to drive mobile proliferation in these tumors. Inactivating PTEN mutations had been prevalent in melanoma (10%), hepatocellular carcinoma (13%), squamous cell carcinoma of the lung (15%), glioblastoma multiforme (29%) and uterine carcinoma (66%) and their presence would lead to eligibility for early research of PI3Kβ inhibition23.
Panorama of scientific actionability
Along with actionable mutations in single genes, different courses of molecular alterations are acknowledged as tumor-agnostic biomarkers of drug response. These embrace mutational profiles brought on by dMMR/POLE mutations and homologous recombination deficiency (HRD), which symbolize phenotypic markers for response to immunotherapy and PARP inhibition respectively. A complete of 319 tumors (3%) exhibited a mutational signature for HRD, which supplies a sign for PARP inhibition remedy and potential sensitivity to platinum chemotherapy24,25,26,27,28. As demonstrated in our companion paper, the etiological foundation of HRD was, nonetheless, solely identifiable in 16% of those instances primarily based on biallelic inactivation of BRCA1, BRCA2, PALB2, BRIP1 or RAD51B by way of germline and somatic mutations29. Whereas different instances could also be brought on by promoter methylation, which couldn’t be assessed as a result of these information will not be out there for 100kGP samples, the findings present a robust rationale for extending the variety of sufferers doubtlessly eligible for PARP inhibitors quite than solely counting on BRCA-testing. A complete of 1,309 tumors possessed a excessive coding tumor mutational burden (greater than ten mutations per megabase, Mb) and 144 cancers had proof of dMMR. Contemplating these collectively would recommend that 1,312 sufferers could also be eligible for checkpoint inhibition30,31. To discover the prospect of a number of focused therapies being utilized in the identical affected person, we mixed the OncoKB scientific actionability annotations with that of TMB, dMMR and HRD scientific actionability annotations. In whole, 11,503 impartial distinctive gene targets had been current in 6,151 samples with 34% (3,577 of 10,478) of tumors possessing one, 13% (1,361 of 10,478) two and 12% (1,213 of 10,478) possessing at the least three clinically actionable driver mutations.
Increasing the druggable most cancers genome
A chance rising from the systematic evaluation of most cancers genomes is the identification of latest therapeutic intervention methods. Of the 330 candidate most cancers driver genes recognized on this research, 261 (79%) will not be presently recognized as therapeutic targets in both COSMIC or OncoKB databases. As a way of triaging these genes as candidates for therapeutic intervention, we assessed the essentiality and selectivity of driver genes and their druggability utilizing RNAi/CRISPR DepMap information and the integrative cancer-focused knowledgebase, canSAR, respectively32,33. We discovered 96 of 261 (37%) of those genes are predicted to be generally important and of those 12 of 96 (13%) have a chemical probe out there and 35 of 96 (36%) have a ligandable three-dimensional (3D) construction (Supplementary Desk 11).
Motivated by the commentary that concentrating on proteins which work together with most cancers driver genes may end up in profitable precision oncology methods, we sought to develop the community of druggable targets in most cancers34,35. To this finish, we used canSAR to map and pharmacologically annotate networks of the most cancers genes recognized for every tumor sort. Particularly, we seeded networks with driver genes recognized in every tumor group and used transcriptional and curated protein–protein interactions to get better a refined cancer-specific community of proteins, every protein being annotated on the idea of a number of assessments of ‘druggability’, that’s the probability of the protein being amenable to small molecule drug intervention. After seeding every cancer-specific community with their respective drivers, we yielded a complete of 631 distinct proteins throughout all cancers (Supplementary Desk 12). The median variety of distinctive proteins in every community throughout all cohorts was 57, with colorectal most cancers possessing the most important community (n = 231; Prolonged Information Fig. 9) and spindle cell carcinoma possessing the smallest community (n = 10). As anticipated there was a correlation between community dimension and variety of recognized drivers for every most cancers sort (Pearson’s r = 0.9, P = 1.23 × 10−9).
Of those 631 proteins, 58% (n = 369) had been retrieved solely by way of community evaluation, of which most (n = 323) weren’t formally recognized as candidate driver genes in any most cancers sort (hereafter known as cancer-network proteins). Notable examples embrace HDAC1, CDK2 and CDK1, which had been current in 31, 29 and 28 cohorts, respectively. We noticed 70% (n = 225) of those cancer-network proteins as being targetable by present accredited or investigational therapies, with notable examples together with BCL2 and BTK. Of the remaining 97 genes, 34 are generally important, 11 possess concordant lineage specificity, 48 are ligandable by 3D construction and 11 have an present high-quality probe out there (Supplementary Desk 13). Collectively these information present potential future alternatives for remedy for a number of cancers. For instance, CDC5L, a core part of the Prp19 (hPrp19)/Cdc5L pre-RNA splicing complicated, is a part of the melanoma most cancers protein community36. This protein is predicted to be generally important with lineage specificity and has a 3D ligandable construction.