Roughly 25–35% of grownup sufferers with acute myeloid leukemia (AML) carries NPM1 mutation, which usually indicated a good final result within the absence of FLT3-ITD mutation [1]. NPM1 mutations are absent in clonal hematopoiesis, and have been thought-about as AML initiating lesions [2]. Analysis on co-mutation traits of NPM1-mutated sufferers targeting FLT3-ITD, which has been recommended to carry a unfavorable prognostic affect on NPM1-mutated sufferers by a number of giant retrospective medical research [3, 4]. In addition to FLT3-ITD, though there stays controversy, different high-frequency co-mutations comparable to DNMT3A, IDH1, IDH2, FLT3-TKD, NRAS, and WT1 mutations have additionally been identified to have an effect on the prognosis of NPM1-mutated sufferers [3, 5,6,7,8,9]. Certainly, identification of particular co-mutation mixtures aside from FLT3-ITD mutation is crucial for exact threat stratification and remedy technique optimization for NPM1-mutated AML sufferers. Since allogeneic hematopoietic stem cell transplantation (allo-HSCT) is mostly thought-about to enhance the long-term final result of most adverse-risk and appropriate intermediate-risk AML sufferers, for NPM1-mutated AML sufferers, it’s crucial to revisit the co-mutation profiles to find out the optimum inhabitants who might profit from allo-HSCT.
On this research, we carried out a retrospective evaluation of newly identified grownup AML sufferers with NPM1 mutations (acute promyelocytic leukemia excluded) in our middle identified from October 2018 to December 2022, specializing in exploring the therapeutic and prognostic significance of co-mutation traits in AML sufferers with NPM1 mutations. Sufferers who obtained no less than one full course of induction remedy had been included within the additional final result evaluation. Desk S1 offered particulars of induction chemotherapy. We evaluated efficacy after two induction cycles, except sufferers achieved CR/CRi after receiving just one induction cycle or discontinued remedy. Response analysis was carried out in line with the NCCN pointers for AML (model 3. 2023) and was categorized as CR/CRi or non-CR/CRi (together with PR and NR) cohort [10]. Total survival (OS) was outlined because the time interval from remedy initiation till dying attributable to any cause. Occasion-free survival (EFS) was outlined because the time interval from remedy initiation to the incidence of induction failure, relapse, or dying, whichever got here first. Illness-free survival (DFS) was outlined because the time interval from illness remission to the incidence of relapse or dying, whichever got here first. The research was carried out in accordance to the Declaration of Helsinki and was authorized by the Ethics Committee of the First Affiliated Hospital of Zhejiang College School of Drugs (Hangzhou, China, Ethics Approval Quantity: IIT20240304A). All statistical analyses had been carried out utilizing GraphPad Prism 7.0 software program (GraphPad Software program, CA, USA) and SPSS 23.0 (SPSS Inc., Chicago, IL).
100 ninety-two newly identified NPM1-mutated AML sufferers detected by way of next-generation sequencing (NGS) had been analyzed (Tables S2–S4). Twenty NPM1 mutants had been recognized, most of which had been positioned in exon 12 and manifested as 4 base pair duplication/insertion alteration. Seven non-exon 12 mutants had been positioned in exon 5, 8, 9 and exon 11, respectively (Fig. 1A and Desk S5). A complete of 56 co-mutated genes had been detected within the cohort (Fig. 1B). Co-mutated genes with a detection fee of ≥10% included FLT3 (56.77%), DNMT3A (48.44%), TET2 (29.69%), IDH2 (23.96%), IDH1 (14.58%), PTPN11 (11.46%), and NRAS (11.46%). Co-mutated genes associated to epigenetics and sign transduction had been the most typical by useful classification (Desk S6).
A Protein area construction and site of amino acids affected by mutations in NPM1. A number of nuclear import and export alerts of NPM1 help its nucleocytoplasmic shuttling and cytological localization. The conserved N-terminal area of NPM1 accommodates a leucine-rich nuclear export sign (NES). The center area accommodates two nuclear localization alerts (NLS) that drive NPM1 to maneuver from the cytoplasm to the nucleus. The C-terminus accommodates a nucleolar localization sign (NoLS), through which two extremely conserved tryptophan residues (W288 and W290) are accountable for the proper folding of the helix to stabilize the hydrophobic core of NoLS. Many of the insertion mutations in exon 12 led to the lack of the unique NoLS sign and generated a brand new NES sign, resulting in aberrant cytoplasmic dislocation of NPM1 protein. B Co-mutation distribution map of NPM1-mutated AML sufferers.
100 seventy-eight sufferers (92.71%) obtained no less than one full course of intensive induction chemotherapy and underwent efficacy evaluation, of which 133 sufferers (74.72%) achieved CR/CRi inside two programs of induction chemotherapy. The median follow-up of the 178 sufferers was 26.23 months (95% confidence interval [CI], 23.31–29.16). The median OS and DFS haven’t been reached, with the median EFS of 15.03 months (95% CI, 8.25–21.82). The three-year anticipated OS, EFS, and DFS had been 51.5%, 40.3%, and 53.7%, respectively.
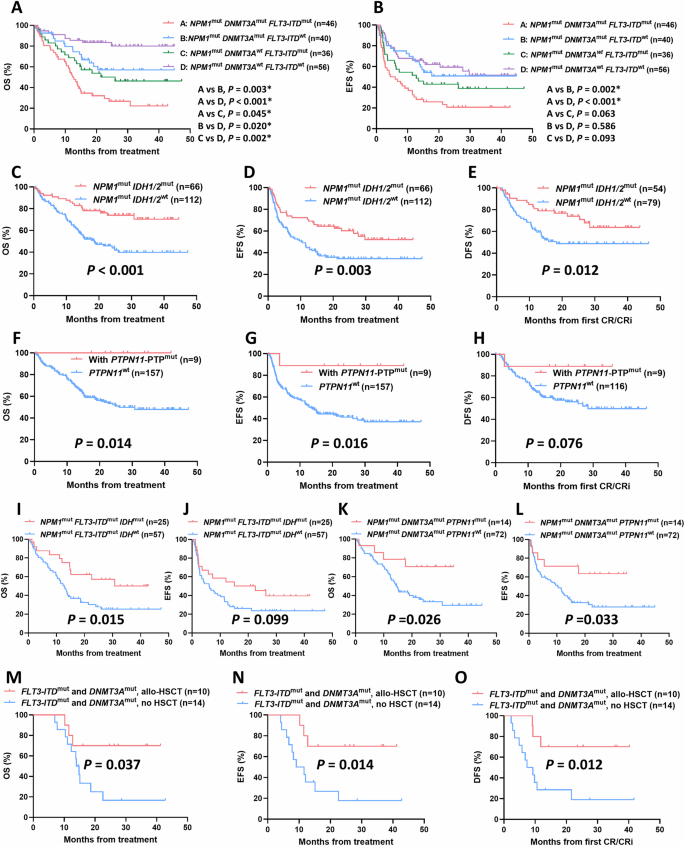
Whatever the cut-off worth of variant allele frequency (VAF) ranges, there was no vital distinction in OS, EFS, and DFS between NPM1low VAF group and NPM1excessive VAF group (Fig. S1). Then we centered on affect of co-mutations on response and final result of AML sufferers with NPM1 mutations. Among the many 178 NPM1-mutated sufferers included within the follow-up, we seen that sufferers with both FLT3-ITD or DNMT3A mutations confirmed considerably worse CR/CRi charges and prognosis developments than wild sort group (FLT3-ITD, CR/CRi charges, 63.41% vs. 84.38%, p = 0.001; median OS, 14.3 months vs. NR, p < 0.001; median EFS, 7.3 months vs. NR, p < 0.001; median DFS, 21.6 months vs. NR, p = 0.044; DNMT3A, CR/CRi charges, 67.44% vs. 81.53%, p = 0.013; median OS, 15.3 months vs. NR, p < 0.001; Median EFS, 11.6 months vs 27.7 months, p = 0.031; Median DFS, p = 0.337) (Desk S7 and Fig. S2). We additional divided sufferers into 4 subgroups in line with the FLT3-ITD and DNMT3A mutation standing. NPM1/FLT3-ITD/DNMT3A triple mutants confirmed extraordinarily poor OS and EFS developments amongst 4 teams (Fig. 2A, B). In addition to, we seen that when mixed with DNMT3A mutations, FLT3-ITD mutated sufferers exhibited considerably worse OS than that of FLT3-ITD wild-type sufferers (p = 0.003), whereas related outcomes had been present in DNMT3A wild-type sufferers (p = 0.002); We additionally seen that when mixed with FLT3-ITD mutations, DNMT3A mutated sufferers exhibited considerably worse OS than that of DNMT3A wild-type sufferers (p = 0.045), with related outcomes occurred in FLT3-ITD wild-type sufferers (p = 0.020) (Fig. 2A).
A OS and B EFS of NPM1-mutated AML sufferers with totally different mixture patterns of FLT3-ITD and DNMT3A mutations. C OS, D EFS, and E DFS of NPM1-mutated AML sufferers with IDH1/2 mutation. F OS, G EFS, and H DFS of NPM1-mutated AML sufferers with PTPN11-PTP mutation. I OS and J EFS of NPM1mutFLT3-ITDmut AML sufferers with IDH mutations. Ok OS and L EFS of NPM1mutDNMT3Amut AML sufferers with PTPN11 mutations. M OS, N EFS, and O DFS of allo-HSCT on NPM1-mutated AML sufferers harbored each FLT3-ITD and DNMT3A mutations.
For sufferers mixed with IDH1/2 mutations, we noticed that the IDH1/2 mutant group considerably improved OS, EFS, and DFS in contrast with wild-type group (Median OS, NR vs. 18.6 months, p < 0.001; Median EFS, NR vs 10.2 months, p = 0.003; Median DFS, NR vs 18.3 months, p = 0.012) (Figs. 2C–E and S3). Though sufferers mixed with PTPN11 mutations confirmed a development towards improved final result in contrast with PTPN11 wild-type, the distinction was not vital (Fig. S4). PTPN11 mutations have been reported to be primarily clustered within the N-terminal Src homology area 2 (N-SH2) and phosphatase (PTP) domains. Since mutations in each two domains concerned in attenuating the autoinhibition of the protein, SHP2, encoded by PTPN11 [11], we additional investigated whether or not mutations in several domains of PTPN11 led to comparable final result. The OS and EFS of sufferers with PTPN11-PTP area mutations had been considerably improved in comparison with these with PTPN11 wild-type (Median OS, NR vs 26.0 months, p = 0.014; Median EFS, NR vs 13.5 months, p = 0.016). Related developments had been present in DFS, whereas sufferers with PTPN11-N-SH2 area mutations confirmed no vital enchancment in final result (Figs. 2F–H and S4). As well as, Fig. S5 confirmed the prognostic affect of different co-mutation genes with a detection fee of ≥10% within the follow-up sufferers, together with TET2, FLT3-TKD, NRAS, and WT1, with developments all non-significant.
Additional, we took into consideration the presence of IDH or PTPN11 mutations in NPM1-mutated sufferers mixed with FLT3-ITD or DNMT3A mutations to discover the prognostic affect of the precise co-mutation interplay patterns. Individually, carrying IDH mutations considerably improved OS and exhibited an improved EFS development in sufferers with NPM1/FLT3-ITD twin mutations (Median OS, 30.8 vs 12.8 months, p = 0.015; Median EFS, 22.6 vs 6.1 months, p = 0.099), however has no vital affect on the result of sufferers with NPM1/DNMT3A mutations (Figs. 2I, J and S6). Equally, carrying PTPN11 mutations considerably improved OS and EFS in sufferers with NPM1/DNMT3A twin mutations (Median OS, NR vs. 14.6 months, p = 0.026; Median EFS, NR vs. 10.2 months, p = 0.033), however has no vital affect on the result of sufferers with NPM1/FLT3-ITD mutations (Figs. 2K, L and S6).
Earlier analysis usually acknowledged that allo-HSCT is useful for FLT3-ITD mutated AML sufferers with out NPM1 mutations. To establish the subgroup of NPM1-mutated AML sufferers prone to profit from allo-HSCT, we explored the prognosis of sufferers who underwent allo-HSCT throughout post-remission after reaching CR/CRi inside two programs of induction. A complete of 32 sufferers obtained allo-HSCT, with one other 4 sufferers relapsed and obtained salvage-HSCT throughout post-remission. For sufferers with NPM1 mutation, receiving allo-HSCT or salvage-HSCT didn’t considerably enhance the result in contrast with non-transplanted sufferers (Fig. S7). For sufferers with NPM1 mutations mixed with both FLT3-ITD or DNMT3A mutation, allo-HSCT confirmed a development towards improved final result, however the distinction was not vital. When additional centered on sufferers with NPM1/FLT3-ITD/DNMT3A triple mutations characterised by poor prognosis, we noticed that allo-HSCT considerably improved the OS, EFS, and DFS of those subgroup (Median OS, NR vs. 14.0 months, p = 0.037; Median EFS, NR vs. 9.1 months, p = 0.014; Median DFS, NR vs. 7.4 months, p = 0.012) (Fig. 2M–O). However, for NPM1-mutated sufferers with wild sort FLT3-ITD and DNMT3A, administration of allo-HSCT confirmed no improved final result (Fig. S7).
Our outcomes indicated that in NPM1-mutated AML, co-mutations of IDH1/2 and PTPN11-PTP area had been correlated with favorable prognosis, whereas FLT3-ITD and DNMT3A co-mutations had been indicative of poor prognosis. Notably, the presence of NPM1/FLT3-ITD/DNMT3A triple mutations is related to exceptionally adversarial OS and EFS developments. A number of research have reported NPM1/FLT3-ITD/DNMT3A, the most typical triple mutation sample in NPM1-mutated sufferers, outlined an AML subgroup with extraordinarily poor prognosis [7, 12], which aligned with our findings. Additional, our outcomes on particular co-mutation mixtures indicated that IDH and PTPN11 co-mutations, respectively, ameliorated the adversarial prognosis of sufferers with NPM1/FLT3-ITD or NPM1/DNMT3A twin mutations, thus two subsets with improved prognosis had been redefined from the unique adverse-prognosis subset of NPM1-mutated AML. In addition to, for sufferers with NPM1/FLT3-ITD twin mutations, allo-HSCT post-first remission has demonstrated a major enhancement in each OS and DFS juxtaposed with the continued administration of chemotherapy alone [13, 14]. Nevertheless, one other giant cohort research on pediatric AML reported reverse outcomes [15]. Our analysis endeavored to establish the optimum inhabitants who might profit from allo-HSCT. The findings underscored the therapeutic potential of allo-HSCT, significantly for AML sufferers with NPM1/FLT3-ITD/DNMT3A triple mutations throughout post-remission.
In abstract, these findings underscored the significance of co-mutation evaluation in NPM1-mutated AML for threat stratification and therapeutic decision-making, suggesting that allo-HSCT could also be a beneficial technique for NPM1-mutated sufferers with particular adversarial co-mutation profiles. However, additional analysis is required to substantiate these findings and discover how these co-mutations work together to diversify the result of NPM1-mutated AML sufferers.