Acute leukemia is essentially the most often identified malignancy in childhood, with acute lymphoblastic leukemia (ALL) comprising roughly 80% of circumstances in youngsters aged 0–18 years, and acute myeloid leukemia (AML) accounting for roughly 15–20% [1]. The early onset (0–10 years) of most childhood acute leukemias and the excessive concordance fee amongst monozygotic twins counsel a prenatal origin of the illness [1]. Certainly, the presence of preleukemic precursors in twine blood (CB) samples or Guthrie blood spots from youngsters who later developed acute leukemia has been demonstrated in a number of research [2, 3]. Nevertheless, this has primarily been demonstrated experimentally for the commonest cytogenetic subtypes, significantly in ALL.
Probably the most prevalent genetic abnormality in infants (as much as 1 yr of age) with AML (iAML) is chromosome 11q23 translocations involving the KMT2A gene. This happens at a better incidence in youngsters than in adults (38% vs 2%), with the best incidence in infants (77%), suggesting that toddler and grownup AMLs are distinct organic entities. Notably, iAML might have an in utero origin [4], as demonstrated by long-established proof of a prenatal origin for MLL rearrangements [5]. Actually, the t(8;21)(q22;q22) AML subgroup has additionally been related to a prenatal origin [3]. Nevertheless, research of sufferers with AML in numerous molecular subgroups have did not detect the corresponding genetic alteration within the respective CB or Guthrie blood spot samples [6], suggesting a much less frequent prenatal origin of AML in comparison with ALL.
The t(7;12)(q36;p13) is a recurrent chromosomal rearrangement uniquely linked to AML. It ranks because the second commonest abnormality in infants with AML, constituting practically one-third of circumstances and related to a dismal prognosis [7], with extraordinarily poor survival charges and ineffective remedy by hematopoietic stem cell transplantation. Occasion-free survival charges (EFS) are 0–14% and general survival (OS) 0–28%. Nevertheless, more moderen research point out extra optimistic outcomes, with 3-year EFS charges of 43% and 3-year OS charges of 100%, albeit with excessive relapse frequencies [8]. The medical manifestation of t(7;12) primarily presents as AML, though it has been identified in just a few circumstances as B-ALL or biphenotypic leukemia [8]. t(7;12)+ blasts typically exhibit a poorly differentiated immunophenotype. Regardless of missing a particular affiliation with a French-American-British (FAB subtype), blasts are generally categorised as M0, M1, or M2 [8].
The breakpoint on chromosome 7 reveals appreciable heterogeneity, impacting the area 7q31-7q36, which is proximal to the motor neuron and pancreas homeobox 1 (MNX1) gene. There are circumstances reporting sufferers with canonical breakpoints and different harbouring non-classical translocations [8, 9]. This area is completely translocated to the by-product chromosome 12 [7]. Concurrently, the breakpoint on chromosome 12 is located at place 12p13, disrupting the ETS variant transcription issue 6 (ETV6) gene in its 5’ area, particularly between exons 1 and three [8]. Notably, a good portion of t(7;12) circumstances is reported together with particular aneusomies. The presence of a number of further copies of chromosomes 8, 19, or 22 has been constantly noticed [7], with trisomy 19 being significantly prevalent in over 70% of circumstances [8]. The concurrent incidence of those numerical anomalies has been steered as a possible issue within the growth of leukemia. Nevertheless, there is no such thing as a clear proof concerning the mechanistic benefit of buying these further chromosomes, though some research suggest that it could result in the overexpression of particular genes [8, 10]. In step with different circumstances of iAMLs, further mutations are rare. A standard function of t(7;12) sufferers is MNX1 overexpression, which has been studied extensively utilizing in vitro and in vivo fashions trying to recapitulate the biology of this illness [11]. MNX1 overexpression doesn’t result in leukemic transformation of twine blood (CB) cells or grownup mouse bone marrow (BM) cells, however impedes erythroid differentiation and encourages mobile senescence [12]. Nevertheless, MNX1 overexpression in fetal liver cells, however not in grownup BM cells, results in leukemic transformation in a retroviral mouse mannequin [13]. MNX1::ETV6, independently of MNX1 overexpression, doesn’t confer self-renewal capability or leukemogenic potential in a mannequin involving transduced fetal liver cells. Solely an in vitro myeloid biased was described in these assays [13]. Thus, the exact position of the MNX1::ETV6 fusion transcript stays a topic of debate.
Right here we current a case of iAML characterised by the t(7;12) translocation, providing proof supporting the neonatal origin of the illness. Moreover, trisomy 19 is recognized as a secondary oncogenic occasion, possible occurring postnatally within the leukemogenesis course of. A 5-months-old boy was identified with AML, whereby 40% of the BM cells have been immature blasts (MPO+, CD34+, CD38+, CD117+, CD13+, CD33+, CD123−/+, CD9−/+, HLADR+) displaying an aberrant expression of the non-myeloid markers CD7, CD2 and CD56. Cytogenetic evaluation by optical genome mapping (OGM) revealed a karyotype 47, XY,t(7;12)(q36;p13),+19, with no further structural or copy quantity variants (SVs and CNVs) (Fig. 1A). Recurrent mutations for FLT3, NPM1, and TP53 genes have been additionally dominated out. We carried out WGS on AutoMACS-separated BM leukemic blasts (CD34 + CD33+) to characterize the breakpoint and genes concerned within the translocation (Fig. 1B). The CD34-CD33- cell inhabitants was used as non-leukemic management cells. The evaluation confirmed the presence of the translocation with breakpoints at positions chr7:156,958,910 (affecting intron 3 of NOM1), and chr12:11,698,832 (inside intron 2 of ETV6) (Fig. 1C). Moreover, we noticed a 35 Kb inversion (chr12:11,719,998-11,754,857) affecting exon 2 of ETV6 and situated 20 Kb from the translocation breakpoint on chromosome 12. Reconstruction of the chimeric chromosome revealed that the fusion resulted in exons 3-8 of ETV6 in opposing transcriptional orientation in relation to exons 4-11 of NOM1, aside from exon 2 of ETV6, which confirmed the identical transcriptional orientation as NOM1 because of an inversion (Fig. 1C). Additional evaluation revealed a really low mutational burden, with 19 substitutions and indels, none of them affecting coding areas (information not proven), and confirmed the presence of trisomy 19, and a subclonal deletion of the brief arm of chromosome 21.
A. Optical Genome Mapping circos plot depicting the recognized translocation within the diagnostic BM pattern. Low confidence and non-somatic alterations will not be displayed. Every circos monitor signifies (from outer ring): corresponding cytoband, hg38 AML function, structural variants (SVs), copy quantity variants (CNVs), variant allele frequency (VAF) segments and translocations. B. Movement cytometry evaluation of analysis BM pattern earlier than and after the AutoMACs separation of CD34 + CD33+ cells for DNA extraction. The purity of CD34 + CD33+ sorted cells was 94%. C. Genetic alterations recognized by WGS and expression quantification of exons by RNA-seq. Prime panels, illustration of ETV6 and NOM1 exons surrounding the breakpoint within the reference genome and within the translocated allele (t(7;12)). Breakpoints for the t(7;12) in addition to for the inversion affecting exon 2 of ETV6 are indicated (cen, centromere). RNA-seq expression information displaying the presence of a cryptic exon (CE) inside intron 1 of ETV6. Dashed pink traces present splicing occasions between exons within the t(7;12) allele. Center panels, quantification of exon expression by RNA-seq displaying elevated expression of NOM1 exons concerned within the translocation. Backside panels, location of annotated regulatory parts from UCSC browser close to the concerned loci. (cCREs, Candidate Cis-Regulatory parts; GH, Enhancers, and promoters from GeneHancer). D. Expression of MNX1 gene in TARGET dataset samples and the analyzed BM analysis pattern (NOM1::ETV6) (TPM transcripts per million).
In parallel, we carried out RNA-seq of the BM blast cells to discover the purposeful penalties of the t(7;12) translocation. Evaluation confirmed the existence of a chimeric gene connecting exon 2 of ETV6 to a cryptic exon situated inside intron 1 of ETV6 and containing a number of splicing acceptor websites, and the three’-end of this cryptic exon was joined to exon 4 of NOM1 (Fig. 1C). Constantly, expression evaluation revealed pronounced overexpression of the exons of NOM1 within the chimeric transcript in comparison with exons 1-3. By phasing single-nucleotide polymorphisms (SNPs) surrounding the breakpoint and SNPs throughout the exons, we recognized robust allele-specific expression. The variant allele frequency (VAF) for the SNP rs11765440, situated throughout the 3’UTR of NOM1 and in section with the translocation, was 95% by RNA-seq, whereas WGS VAF was solely 50% (information not proven). This means the preferential overexpression of the chimeric allele in these cells. In distinction, a SNP in exon 3 of ETV6 phased with the breakpoint confirmed diminished expression of this ETV6 allele (RNA-seq VAF: 10%, WGS VAF: 57%), supporting the notion that the translocation ends in ETV6 silencing.
Regardless of the noticed overexpression of NOM1, the mechanism by which this transcript would possibly contribute to leukemogenesis stays unclear, significantly because the cryptic exon inside intron 1 of ETV6 had many splice acceptor websites, ensuing within the absence of an open studying body for the chimeric transcript. Subsequently, contemplating the bodily proximity of NOM1 to MNX1 ( < 32 Kb), a pivotal participant within the canonical t(7;12) translocation, we studied whether or not the translocation in NOM1 influenced MNX1 expression. RNA-seq evaluation confirmed that MNX1 was extremely expressed on this pattern (176 transcripts per million) and was among the many prime 2% extremely expressed genes. To match its expression with different AML samples with or with out the t(7;12) translocation, we built-in our expression dataset with that of the TARGET [14]. This evaluation revealed that the pattern from our affected person had the best expression of MNX1 relative to the dataset, adopted by three further circumstances from TARGET that have been constructive for t(7;12) (Fig. 1D). These outcomes strongly counsel that, regardless of involving NOM1, t(7;12) has a serious impact on MNX1 expression, because it was beforehand described [8]. Certainly, the translocated intron 1 of ETV6 accommodates quite a few enhancers and regulatory parts which will drive MNX1 overexpression, mimicking the molecular mechanism described for canonical t(7;12) circumstances [11]. Of be aware, a latest examine factors to an enhancer-hijacking occasion activating the MNX1 promoter from the ETV6 locus as a proof for the MNX1 overexpression on this AML subtype [15]. Total, these observations help that MNX1, reasonably than NOM1, is most probably the motive force occasion of the illness, akin to different t(7;12) leukemias [13].
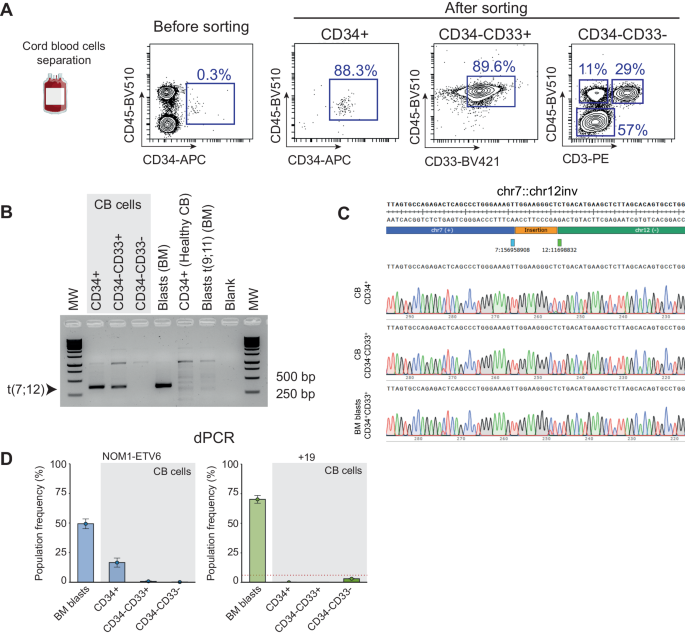
To research the potential neonatal origin of the translocation, we examined the affected person’s cryopreserved CB cells collected at beginning. CB cells separated and enriched for hematopoietic stem and progenitor cells (HSPCs) (CD34+), myeloid progeny (CD34-CD33+) and extra differentiated non-myeloid cells (CD34-CD33-) (Fig. 2A). PCR amplification of the t(7;12) was noticed in each CD34+ HSPCs and the myeloid progeny, however not in differentiated non-myeloid cells (Fig. 2B). The identification of the translocation product was confirmed by Sanger sequencing and was equivalent to that detected by WGS within the analysis BM pattern (Fig. 2C). We then carried out microfluidic digital PCR (dPCR) on DNA from these populations to quantify the proportion of cells with the translocation (Fig. 2D). The t(7;12) translocation could possibly be detected in 18% of CD34+ cells, whereas <1% of CD34-CD33+ and CD34-CD33- cells have been constructive for the translocation. Equally, dPCR was used to guage the copy quantity standing of chromosome 19, the opposite main occasion recognized by OGM and WGS and sometimes related to t(7;12) iAML. Notably, trisomy 19 was under the detection restrict in all CB populations however was readily detected within the analysis BM pattern, indicating that chromosome 19 duplication is a secondary occasion that occurred postnatally. Altogether, our outcomes present the primary proof for the neonatal origin and the cell compartment-of-origin of the translocation t(7;12). In the end, purposeful gain-of-function and or gene-editing experiments with t(7;12)/ETV6::NOM1 fusion in CD34 + HSPC cell fractions might present basic data concerning the leukemogenic potential and exact the cell-of-origin of t(7;12) iAML.
A Movement cytometry evaluation of CB cells earlier than and after AutoMACs separation to isolate the indicated hematopoietic populations for DNA extraction. FACS purity of separated CD34+ HSPCs and CD34-CD33+ myeloid cells was ~90%. Non-myeloid cells have been primarily comprised by erythroblasts (57%), T cells (29%), and non-T cell leukocytes (11%). B PCR amplification of the t(7;12) breakpoint within the totally different hematopoietic populations separated from CB and in BM blasts of the affected person. Unrelated CB cells and a t(9;11) AML cells have been used as controls. C Sanger sequencing of the amplified t(7;12) PCR band from the indicated populations. D dPCR evaluation of the NOM1::ETV6 fusion and trisomy 19 within the totally different indicated populations. Adverse controls have been included to determine the brink (pink line) above which a pattern could possibly be thought of constructive for +19. CB twine blood; BM bone marrow; MW molecular weight.