CelTIL rating and scientific response in early-stage HER2-negative breast most cancers
In SOLTI-1805 TOT-HER3 half A, 78 sufferers with HR+/HER2- breast most cancers obtained a single dose of 6.4 mg/kg HER3-DXd (Fig. 1a). Partly B, a complete of 44 sufferers had been assessed for eligibility (Supplementary Fig. 1a), and 20 sufferers with HR+/HER2- breast most cancers and 17 sufferers with TNBC had been enrolled within the examine to obtain a single dose of 5.6 mg/kg HER3-DXd (Fig. 1a). A compulsory tumor biopsy was carried out at C1D21. The first goal of the trial was to guage modifications within the CelTIL rating, which is a mixed biomarker that integrates the proportion of tumor cellularity and TILs on the identical biopsy11,12,13. Excessive CelTIL scores determine tumors which can be extremely immune infiltrated with lowered tumor cellularity. CelTIL is an early indicator of drug exercise, and probably an early indicator of long-term drug efficacy. Half A demonstrated a rise CelTIL rating after one dose of HER3-DXd, and the principle outcomes had been reported elsewhere7.
a SOLTI-1805 TOT-HER3 trial design and translational examine design. b Consultant examples of H&E staining of a affected person’s tumor with excessive CelTIL response and a affected person’s tumor with low CelTIL response. H&E stainings had been performed for all baseline (n = 114) and day 21 samples (n = 114). c CelTIL rating change in 114 samples of the SOLTI-1805 TOT-HER3 trial half A and half B mixed in all sufferers, in sufferers with scientific response at day 21 (n = 40), and in sufferers with out scientific response at day 21 (C1D21) (n = 56). Pink strains characterize will increase at day 21 whereas blue strains characterize decreases at day 21. P-values (p) had been decided by two-tailed unpaired t checks. Supply information are supplied as a Supply Information file.
Partly B, reported right here for the primary time, the imply age of the examine inhabitants (n = 37) was 51 years (vary, 30–81 years); 20 sufferers (54%) had been premenopausal, and most sufferers introduced with histological grade 2 (30%) or 3 tumors (51%) and invasive ductal carcinoma (89%). The median tumor dimension was 21 mm, with a spread of 10-80 mm. Most sufferers had ERBB3-ultralow tumors (57%). HER3 protein expression was evaluated in 24 sufferers (65%) and most demonstrated excessive total HER3 membrane expression (79%). PAM50 subtype distribution was 46% Basal-like, 24% Luminal A, 19% Luminal B, 11% HER2-enriched (Supplementary Desk 1). The first goal of half B was the rise in CelTIL rating after one dose of HER3-DXd. Total, a imply enhance in CelTIL rating of 9.4 (p = 0.046) was detected (Supplementary Fig. 1b); with imply variations of two.2 and 17.9 in HR+/HER2- and TNBC, respectively. Of the 37 sufferers, 34 had been assessable for scientific response. The scientific ORR was 32% (30% in HR+/HER2− and 35% in TNBC). A big enhance in CelTIL rating (imply enhance = 23.9, p = 0.043) was detected in sufferers with scientific response (Supplementary Fig. 1b). Secondary targets of half B included the affiliation of ERBB3 mRNA with CelTIL at day 21 and security. ERBB3 mRNA expression was not related to CelTIL variation (Supplementary Fig. 1c). Moreover, security partially B was much like that noticed partially A (Supplementary Desk 2), with decrease incidence of hematological and hepatic toxicity in comparison with Half A. No interstitial lung illness nor grade 4/5 occasions had been noticed (Supplementary Desk 3).
To supply additional proof of the affiliation of C1D21 CelTIL with drug efficacy, we mixed SOLTI-1805 TOT-HER3 components A and B, and noticed {that a} constant CelTIL enhance was considerably (p<0.001) related to scientific response (Fig. 1c). Moreover, in SOLTI-1007 NeoEribulin trial14, a rise of CelTIL after 1 cycle of eribulin monotherapy (i.e., at cycle 2 day 1) was considerably related to residual most cancers burden 0 or 1 (RCB-0/1) at surgical procedure after the completion of 4 cycles in sufferers with HER2-negative breast most cancers (Supplementary Fig. 2a, b). Total, these outcomes counsel that CelTIL in HER2-negative breast most cancers is an early readout of drug exercise, and probably an early indicator of drug efficacy and long-term profit.
SOLTI-1805 TOT-HER3 translational examine design
Within the exploratory analyses of SOLTI-1805 TOT-HER3, RNA was remoted from 77 baseline pre-treatment formalin-fixed paraffin-embedded (FFPE) from half A of SOLTI-1805 TOT-HER3, and from 20 FFPE tumor samples from half B for validation functions. DNA was obtained from 49 baseline pre-treatment FFPE tumor samples from half A. Affected person traits are summarized in Supplementary Desk 4. To determine biomarkers of early response to HER3-DXd, we evaluated the affiliation between every baseline variable and CelTIL response (Fig. 1), outlined as an absolute enhance of CelTIL of ≥20 factors between the 2 time factors (i.e., C1D21 minus baseline). Partly A, an absolute enhance of CelTIL of ≥20 factors predicted scientific response with an AUC of 0.68 (p = 0.009) (Supplementary Fig. 3).
Correlative evaluation of CelTIL response and mRNA expression
After a single dose of HER3-DXd, 51 (66%) and 26 (34%) tumors had a high and low CelTIL response, respectively. At baseline, the expression of 41 of 185 (22.2%) genes was discovered related to CelTIL response utilizing SAM evaluation with FDR<10% (Fig. 2a). Of the 41 genes, 37 (90.2%) had been additionally discovered related to CelTIL response utilizing univariate logistic regressions. Total, excessive expression of proliferation and cell division-related genes corresponding to AURKA (p = 0.002), CCNE1 (p = 0.014), and MKI67 (p = 0.010) was related to CelTIL response. Conversely, excessive expression of luminal-related genes corresponding to NAT1 (p = 0.019), SLC39A6 (p = 0.033), MAGED2 (p = 0.021) and THSD4 (p = 0.029) was related to an absence of CelTIL response (Fig. 2b).
a Unsupervised hierarchical clustering of 41 genes had been related to CelTIL response after one dose of HER3-DXd in an unpaired SAM evaluation (FDR<10%). b Forest plot of 37 of genes considerably related to CelTIL response after one dose of HER3-DXd in a logistic regression evaluation in sufferers with CelTIL response (n = 26) and with out CelTIL response (n = 51). Information are introduced as the chances ratios (OR) with error bars displaying 95% confidence intervals. Supply information are supplied as a Supply Information file.
At baseline, 9 of 12 (75.0%) gene expression signatures had been discovered related to CelTIL response. Excessive scores of the PAM50 Basal-like (p = 0.013), PAM50 HER2-enriched (p = 0.044), PAM50 Proliferation (p = 0.002), PAM50 danger of recurrence (ROR) (p = 0.002) and HER2DX Proliferation (p = 0.031) signatures had been related to CelTIL response. Excessive scores of the PAM50 Luminal A (p = 0.001), PAM50 Regular-like (p = 0.036), PAM50 chemo-endocrine delicate (CES) (p = 0.003) and HER2DX HER2 amplicon (p = 0.005) signatures had been related to lack of CelTIL response (Supplementary Fig. 4). Of word, although the Luminal A and the Basal-like signatures had been related to CelTIL response, estrogen receptor (ER) and progesterone receptor (PR) protein expression weren’t (Supplementary Fig. 5).
ERBB2/HER2 expression and HER3-DXd response
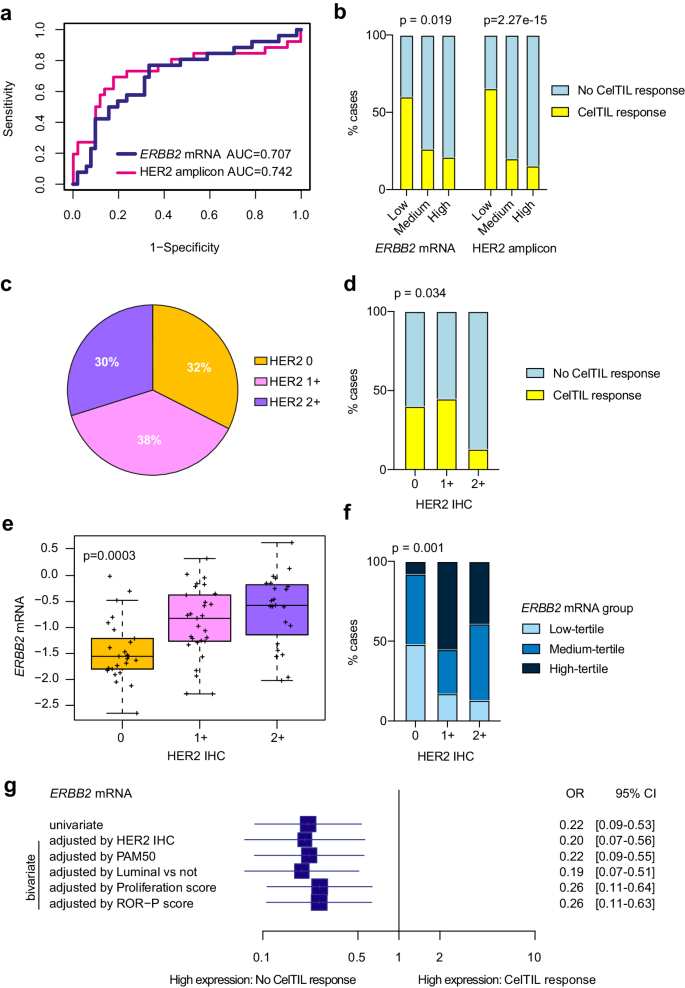
Our pre-treatment RNA-based evaluation recognized ERBB2 mRNA and HER2 amplicon signature as determinants of early response to HER3-DXd. Particularly, low expression of HER2DX ERBB2 and HER2DX HER2 amplicon signature was discovered statistically considerably related to CelTIL response as a steady variable (p = 0.006 and p = 0.005, respectively) (Fig. 3a). We additionally outlined HER2DX ERBB2 and HER2DX HER2 amplicon signature group classes based mostly on the tertiles of the TOT-HER3 half A (Supplementary Fig. 6). HER2DX ERBB2 and HER2DX HER2 amplicon signature group classes outlined by tertiles had been discovered statistically considerably related to lack of CelTIL response (p = 0.019 and p<0.001, respectively), with the proportion of sufferers with a CelTIL response in HER2DX ERBB2 mRNA low-tertile, medium-tertile and high-tertile teams being 60.0%, 26.3% and 21.1%, respectively (Fig. 3b). The distribution of HER2 IHC standing within the 77 baseline tumor samples analyzed was 32% HER2 0+, 38% HER2 1+ and 30% HER2 2+ (Fig. 3c).
a Efficiency of the HER2DX ERBB2 mRNA and HER2 amplicon signature rating to foretell CelTIL response after one dose of HER3-DXd. ROC AUC values are reported. b Proportion of tumors with excessive and low CelTIL response after one dose of HER3-DXd in every HER2DX ERBB2 group (as outlined by tertiles: low n = 26, medium n = 26, excessive n = 25) and every HER2 amplicon signature group (as outlined by tertiles low n = 26, medium n = 26, excessive n = 25). P-values (p) had been decided by two-sided Fisher’s precise check. c Distribution of HER2 IHC standing (HER2 0 n = 25, HER2 1+ n = 29, HER2 2+ n = 23) within the 77 baseline samples analyzed. d Proportion of tumors with excessive and low CelTIL response after one dose of HER3-DXd in every HER2 IHC group (HER2 0 n = 25, HER2 1+ n = 29, HER2 2+ n = 23). P-value (p) was decided by two-sided Fisher’s precise check. e Boxplot displaying HER2DX ERBB2 mRNA expression throughout HER2 IHC teams (HER2 0 n = 25, HER2 1+ n = 29, HER2 2+ n = 23). For the boxplot, middle line signifies median; field limits point out higher and decrease quartiles; whiskers point out 1.5× interquartile vary. P-value (p) was decided by one-way ANOVA. f Proportion of tumors with excessive, medium and low HER2DX ERBB2 mRNA (as outlined by tertiles: low n = 26, medium n = 26, excessive n = 25) in every HER2 IHC group. P-value (p) was decided by two-sided Fisher’s precise check. g Forest plot of the affiliation of HER2DX ERBB2 mRNA with CelTIL response in univariate and bivariate logistic regression analyses in sufferers with CelTIL response (n = 26) and with out CelTIL response (n = 51). Information are introduced as the chances ratios (OR) with error bars displaying 95% confidence intervals. Supply information are supplied as a Supply Information file.
Much like HER2DX ERBB2 expression, HER2 protein ranges by IHC had been discovered inversely related to CelTIL response (HER2 0 vs HER2 2+, p = 0.043) and HER2 2+ had a decrease proportion of sufferers with CelTIL response (13.0%) in comparison with HER2 0 (40.0%) and 1+ (45.0%) (p = 0.034) (Fig. 3d). As anticipated, HER2DX ERBB2 mRNA was discovered differentially expressed throughout HER2 0, 1+ and a pair of+ circumstances (Fig. 3e, f). HER2DX ERBB2 mRNA was not related to any PAM50 subtype partially A (Supplementary Fig. 7a). HER2DX ERBB2 mRNA was reasonably correlated with ERBB3 mRNA however no vital correlation was noticed between HER2DX ERBB2 mRNA or HER2 protein ranges by IHC and HER3 protein ranges by IHC (Supplementary Fig. 7b, c). A sensitivity evaluation utilizing completely different cutoffs of CelTIL response (>0, ≥10, ≥20, ≥30, ≥40) was carried out. HER2DX ERBB2 mRNA ranges had been inversely related to CelTIL response in all explored cutoffs, with the very best affiliation at CelTIL response cutoff ≥20 (Supplementary Fig. 8).
Subsequent, we evaluated the flexibility of HER2DX ERBB2 expression to foretell CelTIL response independently of different molecular variables. Throughout numerous logistic regression bivariate analyses, greater HER2DX ERBB2 mRNA expression was considerably related to decrease CelTIL response after adjusting for HER2 IHC (p = 0.002), PAM50 subtype (p = 0.001), PAM50 luminal vs non-luminal (p = 0.001), PAM50 proliferation (p = 0.003) and PAM50 ROR (p = 0.003) (Fig. 3g). Curiously, the affiliation of HER2 IHC with CelTIL response was misplaced (p = 0.538) when HER2DX ERBB2 expression was included within the logistic regression mannequin.
Lastly, we examined the flexibility of HER2DX ERBB2 mRNA expression to foretell CelTIL response in an impartial dataset of 20 sufferers with early-stage HR+/HER2− breast most cancers handled in Half B of the trial. HER2DX ERBB2 expression as a steady variable was discovered statistically considerably related to no CelTIL response (AUC = 0.783) (Supplementary Fig. 9a). Based on the HER2DX ERBB2 tertiles recognized partially A, the proportion of sufferers with a CelTIL response throughout HER2DX ERBB2 low-tertile, medium-tertile and high-tertile teams recognized partially B was 40%, 29%, and 0%, respectively (Supplementary Fig. 9b).
We subsequent sought to grasp the affiliation between ERBB2 expression and response to chemotherapy in two cohorts of sufferers with HER2-negative breast most cancers (i.e., a neoadjuvant taxane-anthracycline-based cohort, hereafter GSE2506615, and the neoadjuvant eribulin SOLTI-1007-NeoEribulin cohort14). ERBB2 mRNA was not related to response (i.e., pCR or CelTIL response) to chemotherapy (Supplementary Fig. 10).
Lastly, as a further validation of ERBB2 as a possible mechanism of resistance to HER3-DXd, we assessed HER2DX ERBB2 within the baseline tumors of a cohort of 30 breast most cancers patient-derived xenografts (PDXs) handled with HER3-DXd16. Ninety % of PDXs had skilled an entire response or partial response however 53% had a relapse16. PDXs had been labeled as relapsed vs non-relapsed. Though the measurement of resistance was very completely different to that of the SOLTI-1805 TOT-HER3 examine, HER2DX ERBB2 was considerably greater in baseline tumors of PDXs that relapsed in contrast to those who didn’t relapse, additional indicating in the direction of a job of HER2DX ERBB2 within the response of this ADC (Supplementary Fig. 11).
DNA somatic mutations and CelTIL response
Forty-nine of 77 sufferers (64%) in SOLTI-1805 TOT-HER3 half A had tumor DNA-sequencing information obtainable. Baseline traits of the 49 sufferers are summarized in Supplementary Desk 1. CelTIL modifications between the RNA (n = 77) and DNA (n = 49) cohorts had been comparable. All sufferers had at the very least 1 gene somatically mutated. Probably the most often mutated genes had been PIK3CA (33%), GATA3 (29%), ATM (24%), CDH1 (20%), TP53 (18%), KMT2C (16%), KMT2D (16%), and MAP3K1 (14%). The frequency of TP53 mutations was numerically greater (i.e., 100.0% vs. 10.7%) in Basal-like tumors (n = 3/3) in comparison with Luminal A tumors (n = 3/28) (Supplementary Fig. 12a). Among the many most often mutated genes, CelTIL response price was greater in TP53 mutated tumors in comparison with TP53 wild-type tumors (66.7% vs. 25.0%; Fisher’s precise check p = 0.043) (Supplementary Fig. 12b). In logistic regression analyses, TP53 mutations had been considerably related to CelTIL response in univariate evaluation (odds ratio = 6.00, p = 0.024) however this significance was misplaced after adjusting by PAM50 subtype (p = 0.068), proliferation (p = 0.103) and ROR scores (p = 0.110) in all probability as a result of small pattern dimension. Of word, 4 of 49 tumors (8.2%) had mutations in ERBB2, and this alteration was not discovered related to CelTIL response (p = 0.993).
DNA copy-number (CN)-based signatures and CelTIL response
Subsequent, we recognized 150 DNA CN-based signatures beforehand outlined to seize RNA- and protein-based phenotypes such because the PAM50-related biology17,18. A complete of 90 of 150 (60.0%) DNADX signatures had been considerably related to CelTIL response (FDR<10% and univariate logistic regression) (Fig. 4a and Supplementary Fig. 13). Concordant with the gene expression information, excessive scores of signatures associated to the Basal-like biology (e.g., UNC_BASAL_Cluster_Median) and excessive proliferation (e.g., UNC_Scorr_IIE_Correlation and UNC_RB_LOH_Median) had been related to CelTIL response, whereas excessive scores of signatures associated to the Luminal A biology (e.g., UNC_Scorr_IE_Correlation and UNC_Scorr_LumA_Correlation) and HER2 (e.g., HER2 expression and UNC_HER2_Amplicon_Median) had been related to lack of CelTIL response (Fig. 4b).
a Unsupervised hierarchical clustering of 150 CN-based signatures related to CelTIL response after one dose of HER3-DXd in an unpaired SAM evaluation (FDR<10%). b Forest plot of a number of CN-based signatures considerably related to CelTIL response after one dose of HER3-DXd in a logistic regression evaluation in sufferers with CelTIL response (n = 16) and with out CelTIL response (n = 33). Information are introduced as the chances ratios (OR) with error bars displaying 95% confidence intervals. Supply information are supplied as a Supply Information file.
Throughout these 49 sufferers with DNA-sequencing information, HER2DX ERBB2 expression was discovered reasonably correlated with the copy-number sign of chromosome 17q12, the place ERBB2 gene is situated (Pearson correlation coefficient = 0.55, p < 0.001) (Supplementary Fig. 14a). Concordant with this discovering, excessive copy-number sign of 17q12 was additionally discovered inversely related to CelTIL response as a steady variable (p = 0.002; AUC = 0.797) and as group classes outlined by tertiles (p = 0.007; Supplementary Fig. 14b). For instance, the proportion of sufferers with a CelTIL response in 17q12 CN section low-tertile, medium-tertile and high-tertile teams was 56.3%, 35.3% and 6.3%, respectively (Supplementary Fig. 14c). Lastly, we evaluated the efficiency of the two CN-based HER2-related signature scores as steady variables to foretell CelTIL response after one dose of HER3-DXd. The ROC AUC values for the CN-based HER2-IHC signature (which predicts HER2 protein expression) and the CN-based HER2 amplicon signature (which predicts ERBB2 amplifications) had been 0.812 and 0.843, respectively (Supplementary Fig. 14b). As categorical variables outlined by tertiles, the CN-based HER2-IHC signature and the CN-based HER2 amplicon signature had been additionally considerably related to CelTIL response. CelTIL response in CN-based HER2-IHC signature-low, -medium, and -high teams had been 59%, 25%, and 12%, respectively. CelTIL response in CN-based HER2 amplicon signature-low, -medium and -high teams had been 65%, 19%, and 12%, respectively (Supplementary Fig. 14c).
CN-based molecular subtypes and CelTIL response
Utilizing unsupervised evaluation from 150 CN-based signatures, we now have not too long ago recognized 4 DNADX subtypes (i.e., clusters -1, -2, -3, and -4) with prognostic worth inside HR+/HER2- breast most cancers18. Throughout 49 tumors from SOLTI-1805 TOT-HER3 trial half A with DNA-sequencing information, the proportion of sufferers with DNADX cluster-1 illness was 34.7%, cluster-2 was 30.6%, cluster-3 was 22.5%, and cluster-4, 12.2% (Fig. 5a). As anticipated, DNADX cluster-1 had excessive scores of Luminal A-like signatures, DNADX cluster-2 had excessive scores of estrogen-related and Luminal B-like signatures, DNADX cluster-3 had excessive scores of the Basal-like biology and proliferation-related signatures, and DNADX cluster-4 had excessive scores of HER2, Luminal B-like and proliferation signatures (Fig. 5b). Concordantly, all PAM50 Basal-like tumors (n = 3) had been recognized as being a part of DNADX cluster-3 (Fig. 5c), and no PAM50 ROR-low tumor (n = 5) was recognized as being a part of DNADX cluster-3 (Fig. 5d). To supply some extra perception into the biology of the 4 DNADX subtypes, we assessed the distribution of the DNADX subtypes throughout the integrative clusters (IntClust 1-10) of the METABRIC cohort19. There have been vital variations within the distribution of DNA subtypes (clusters 1-4) (chi-square p < 0.001). Greater illustration of DNA-based clusters 3 and 4 had been noticed within the integrative clusters of worse prognosis (i.e: IntClust 5, 9, and 10), whereas the DNA-based clusters 1 and a pair of had been extra recognized in these integrative clusters of higher prognosis (i.e. IntClust 3, 4, 7, 8) (Supplementary Fig. 15).
a Distribution of DNADX CN-based subtypes in 49 tumors of the TOT-HER3 trial (Cluster 1 n = 17, Cluster 2 n = 15, Cluster 3 n = 11, Cluster 4 n = 6). b Unsupervised hierarchical clustering of 150 CN-based signatures in every CN-based subtype. The correlation (Cor) values of signatures monitoring related phenotypes are reported close to the dendogram. c Proportion of PAM50 molecular subtypes in every DNADX CN-based subtype (Cluster 1 n = 17, Cluster 2 n = 15, Cluster 3 n = 11, Cluster 4 n = 6). d Proportion of PAM50 ROR excessive, medium and low rating in every DNADX CN-based subtype (Cluster 1 n = 17, Cluster 2 n = 15, Cluster 3 n = 11, Cluster 4 n = 6). e Proportion of excessive and low CelTIL responders after one dose of HER3-DXd in every DNADX CN-based subtype (Cluster 1 n = 17, Cluster 2 n = 15, Cluster 3 n = 11, Cluster 4 n = 6). P-values (p) in (c–e) had been decided by two-sided Fisher’s precise check. Supply information are supplied as a Supply Information file.
Lastly, we evaluated the affiliation of the 4 DNADX CN-based subtypes with CelTIL response. Total, the 4 DNADX subtypes had been discovered related to CelTIL response (p < 0.001). The proportion of sufferers with CelTIL response throughout the 4 DNADX subtypes was 90.9% (cluster-3), 33.3% (cluster-4), 17.6% cluster-1, and 6.7% cluster-2 (Fig. 5e). Cluster-3 was discovered considerably related to CelTIL response in comparison with clusters 1-2-4 (84.2% vs. 9.1%; odds ratio = 53.33, p < 0.001) and this affiliation was discovered impartial of HER2DX ERBB2 mRNA expression (odds ratio = 20.13, p = 0.040). On this bivariate evaluation, HER2DX ERBB2 mRNA was additionally impartial of DNADX cluster 3 (odds ratio = 0.04, p = 0.008).