Sufferers
Of the 802 sufferers with RAS WT mCRC included within the PARADIGM efficacy evaluation inhabitants, 733 sufferers (91.4%) supplied knowledgeable consent for this biomarker research and had baseline blood plasma samples that have been evaluable for ctDNA (Fig. 1). Amongst these 733 sufferers, 554 sufferers (75.6%) had left-sided main tumors, 169 (23.1%) had right-sided main tumors, and 10 (1.4%) had a number of main lesions in each the left and proper sides. For the biomarker-evaluable inhabitants, median follow-up as of the information cutoff date (14 January 2022) was 61.4 months (95% confidence interval (CI), 60.5–62.9 months) within the panitumumab + mFOLFOX6 group and 60.5 months (95% CI, 59.5–62.9 months) within the bevacizumab + mFOLFOX6 group.
a‘Adverse hyperselected’ was outlined as plasma ctDNA being detrimental for all prespecified gene alterations, together with mutations in BRAF V600E, KRAS, PTEN, EGFR ECD exons 1–16 and NRAS, amplifications of HER2 and MET, and gene fusions of RET, NRTK1 and ALK. b‘Gene altered’ was outlined as detection of any of the next in ctDNA: a mutation in BRAF V600E, KRAS, PTEN, EGFR ECD exons 1–16 and/or NRAS, amplification of HER2 and/or MET, and gene fusion of RET, NRTK1 and/or ALK. cSome sufferers had a number of main lesions on each the left and proper sides. The dotted line represents a further exploratory evaluation assessing genetic alterations of MSS/MSI standing and RAS/BRAF mutations based mostly on guideline suggestions. ECD, extracellular area; QC, high quality management.
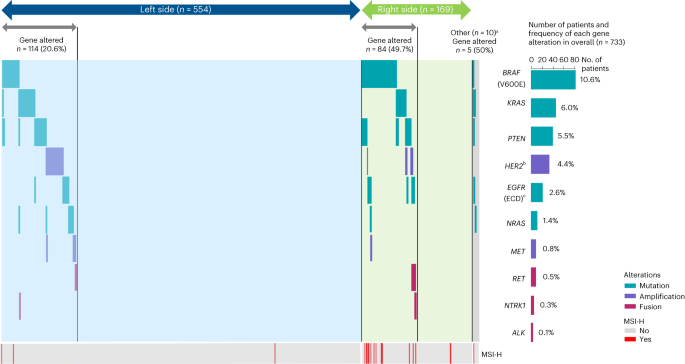
Affected person ctDNA was assessed for 90 mutations, 26 amplifications and three rearrangements in mCRC-related genes utilizing a customized NGS-based panel (Strategies). Most variant allele frequency is reported for all samples in Supplementary Desk 1. We report outcomes of a preplanned evaluation for detrimental hyperselection, which means plasma ctDNA was detrimental for all prespecified gene alterations related to resistance to anti-EGFR antibody remedy15,17,22,23,24, together with mutations in BRAF V600E, KRAS, NRAS, PTEN and EGFR ECD exons 1–16, amplifications of HER2 and MET, and gene fusions of RET, NRTK1 and ALK. A complete of 530 sufferers (72.3%) met these detrimental hyperselection standards (Desk 1). Sufferers with left-sided main tumors met detrimental hyperselection standards at a better charge (79.4%: 440 of 554 sufferers) than sufferers with right-sided main tumors (50.3%: 85 of 169 sufferers).
Among the many 203 (27.7%) sufferers with at the very least one gene alteration, the most typical alterations have been BRAF V600E mutation (10.6%), KRAS mutation (6.0%) and PTEN mutation (5.5%) (Fig. 2). Most sufferers had just one mutation within the left-sided (93.0%; 106 of 114 sufferers), right-sided (73.8%; 62 of 84 sufferers) and general populations (84.2%; 171 of 203 sufferers), with co-occurrence of a number of mutations most typical in right-sided mCRC (Fig. 2). The frequency of gene alterations is summarized by main tumor location and remedy group in Prolonged Information Desk 1.
Outcomes by detrimental hyperselection standing
Total survival
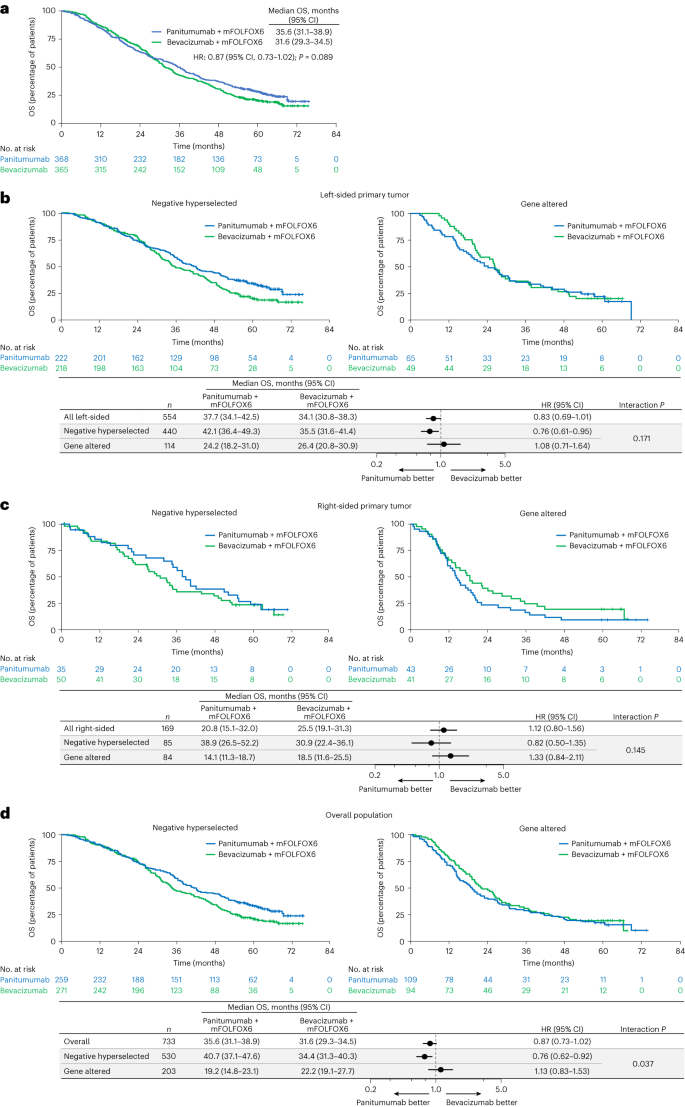
For the whole biomarker-evaluable inhabitants, median OS was 35.6 months (95% CI, 31.1–38.9 months) with panitumumab + mFOLFOX6 and 31.6 months (95% CI, 29.3–34.5 months) with bevacizumab + mFOLFOX6 (HR for demise stratified by age and presence of liver metastasis: 0.87; 95% CI, 0.73–1.02; Fig. 3a). For sufferers assembly detrimental hyperselection standards (that’s, no gene alteration detected), OS was longer with panitumumab versus bevacizumab in sufferers with left-sided main tumors (median 42.1 versus 35.5 months; HR, 0.76; 95% CI, 0.61–0.95; P worth for interplay between remedy group and detrimental hyperselection standing = 0.171; Fig. 3b), and there was a development for longer OS with panitumumab versus bevacizumab in sufferers with right-sided tumors (38.9 versus 30.9 months; HR, 0.82; 95% CI, 0.50–1.35; interplay P = 0.145; Fig. 3c). Within the general detrimental hyperselected inhabitants, median OS was longer with panitumumab versus bevacizumab (40.7 versus 34.4 months; HR, 0.76; 95% CI, 0.62–0.92; interplay P = 0.037; Fig. 3d).
a, Kaplan–Meier estimates of OS within the general biomarker-evaluable inhabitants (all ctDNA-evaluable sufferers). b–d Kaplan–Meier estimates of OS by detrimental hyperselection standing in sufferers with left-sided main tumors (b), sufferers with right-sided main tumors (c) and the general inhabitants (all ctDNA-evaluable sufferers) (d). The forest plots under the Kaplan–Meier plots in b, c and d present HR ± 95% CI. A Cox proportional hazard mannequin with out stratification elements was used to calculate HRs for group comparisons and P values for the interplay between detrimental hyperselection standing and remedy group. Statistical checks have been two-sided with out adjustment for a number of comparisons.
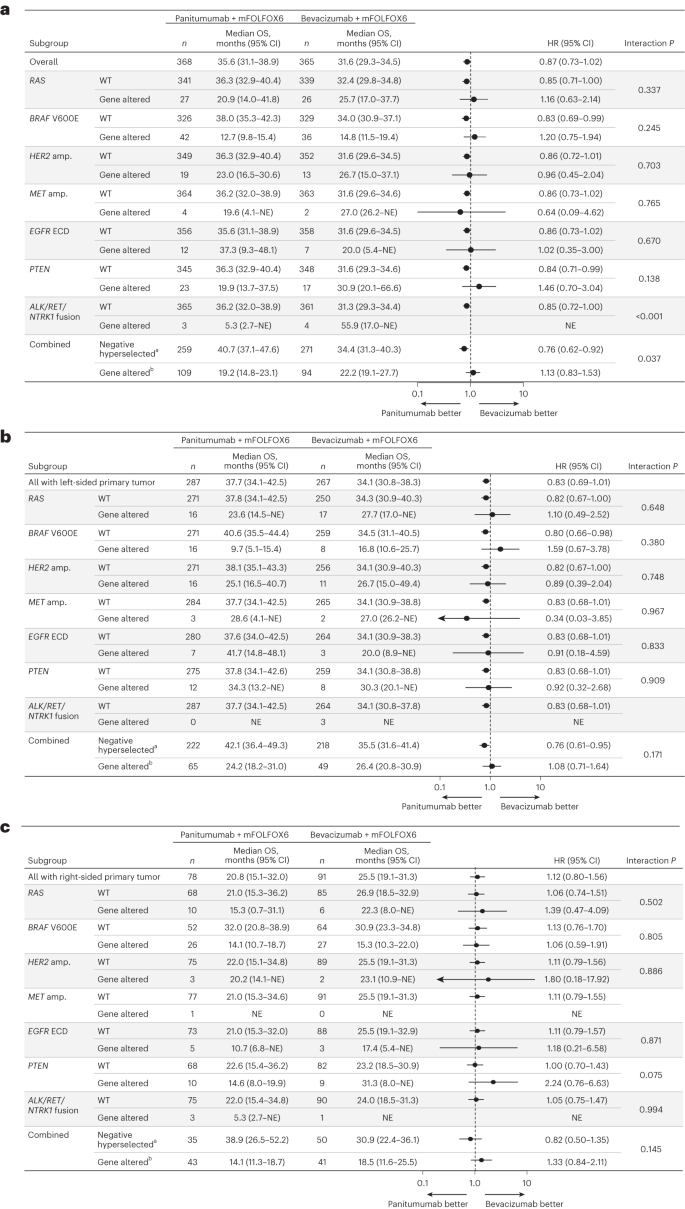
For sufferers with any gene alteration, median OS was related or inferior with panitumumab versus bevacizumab no matter main tumor sidedness. Median OS with panitumumab versus bevacizumab in gene-altered sufferers was 24.2 versus 26.4 months in sufferers with left-sided main tumors (HR, 1.08; 95% CI, 0.71–1.64; Fig. 3b), 14.1 versus 18.5 months in sufferers with right-sided main tumors (HR, 1.33; 95% CI, 0.84–2.11; Fig. 3c) and 19.2 versus 22.2 months within the general inhabitants (HR, 1.13; 95% CI, 0.83–1.53; Fig. 3d). Outcomes of the subgroup evaluation of OS by particular gene alterations are proven for the general inhabitants in Fig. 4a, and for the left-sided and right-sided populations in Fig. 4b and 4c, respectively.
a–c, OS by particular gene alteration within the general inhabitants (a), sufferers with left-sided main tumors (b) and sufferers with right-sided main tumors (c). Information plotted are HRs ± 95% CI. A Cox proportional hazard mannequin with out stratification elements was used to calculate HRs for group comparisons and P values for the interplay between detrimental hyperselection standing and remedy group. Statistical checks have been two-sided with out adjustment for a number of comparisons. aAdverse hyperselected sufferers have been WT for all the following: RAS, BRAF V600E, HER2 amp., MET amp., EGFR ECD, PTEN and ALK/RET/NTRK1 fusion. bGene-altered sufferers had at the very least one of many following alterations: RAS, BRAF V600E, HER2 amp., MET amp., EGFR ECD, PTEN or ALK/RET/NTRK1 fusion. amp., amplification; NE, not estimable.
Development-free survival
For detrimental hyperselected sufferers, progression-free survival (PFS) was related with panitumumab + mFOLFOX6 versus bevacizumab + mFOLFOX6 within the left-sided (14.0 versus 12.8 months; HR, 0.91; 95% CI, 0.73–1.13; P worth for interplay between remedy group and detrimental hyperselection standing = 0.049), right-sided (13.2 versus 11.3 months; HR, 1.08; 95% CI, 0.66–1.77; interplay P = 0.025) and general populations (median, 13.6 versus 12.8 months; HR, 0.92; 95% CI, 0.75–1.12; interplay P < 0.001; Prolonged Information Fig. 1). For sufferers with any gene alteration, median PFS was related with panitumumab and bevacizumab within the left-sided inhabitants (9.3 versus 9.9 months; HR, 1.45; 95% CI, 0.94–2.23) however shorter with panitumumab than bevacizumab within the right-sided (6.3 versus 10.3 months; HR, 2.25; 95% CI, 1.36–3.70) and general populations (7.8 versus 9.8 months; HR, 1.68; 95% CI, 1.23–2.29; Prolonged Information Fig. 1).
Response charge
Amongst detrimental hyperselected sufferers, response charges have been larger with panitumumab versus bevacizumab within the left-sided inhabitants (83.3% (95% CI, 77.8–88.0) versus 66.5% (95% CI, 59.8–72.7); odds ratio (OR), 2.52 (95% CI, 1.61–3.98); interplay P = 0.012), with an analogous development within the right-sided inhabitants (71.4% (95% CI, 53.7–85.4) versus 66.0% (95% CI, 51.2–78.8); OR, 1.29 (95% CI, 0.51–3.37); interplay P = 0.060; Prolonged Information Fig. 2), though the right-sided between-group distinction was comparatively small (+5.4%). Within the general detrimental hyperselected inhabitants, the response charge was larger with panitumumab (81.5% (95% CI, 76.2–86.0)) than with bevacizumab (66.8% (95% CI, 60.8–72.4)); OR, 2.19 (95% CI, 1.47–3.29); interplay P < 0.001). For sufferers with any gene alteration, the response charge was related with panitumumab (67.7% (95% CI, 54.9–78.8)) versus bevacizumab (73.5% (95% CI, 58.9–85.1); OR, 0.76 (95% CI, 0.33–1.70)) within the left-sided inhabitants however decrease with panitumumab (41.9% (95% CI, 27.0–57.9)) than bevacizumab (65.9% (95% CI, 49.4–79.9); OR, 0.37 (95% CI, 0.15–0.89)) within the right-sided inhabitants, with an analogous development within the general gene-altered inhabitants (57.8% (95% CI, 48.0–67.2) versus 69.1% (95% CI, 58.8–78.3); OR, 0.61 (95% CI, 0.34–1.09); Prolonged Information Fig. 2).
Depth of response
Median depth of response (most change in goal lesion measurement) was higher with panitumumab versus bevacizumab amongst detrimental hyperselected sufferers with left-sided tumors (−60.2% (95% CI, −64.0 to −58.8) versus −43.6% (95% CI, −47.9 to −39.4)) and right-sided tumors (−56.4% (95% CI, −67.7 to −51.3) versus −39.4% (95% CI, −52.7 to −31.3)) and within the general detrimental hyperselected inhabitants (−60.2% (95% CI, −63.8 to −57.6) versus −43.6% (95% CI, −47.4 to −39.4)). In gene-altered sufferers, depth of response was related with panitumumab and bevacizumab within the left-sided (−53.6% (95% CI, −60.7 to −46.0) versus −44.2% (95% CI, −48.8 to −35.1)), right-sided (−30.0% (95% CI, −42.1 to −9.8) versus −53.3% (95% CI, −61.1 to −35.8)) and general populations (−46.0% (95% CI, −53.3 to −33.4) versus −45.1% (95% CI, −52.3 to −37.9); Prolonged Information Fig. 3).
Healing resection charge
For detrimental hyperselected sufferers, the healing resection charge was larger with panitumumab versus bevacizumab within the left-sided inhabitants (19.8% (95% CI, 14.8–25.7) versus 10.6% (95% CI, 6.8–15.4); OR, 2.10 (95% CI, 1.23–3.66)) and related between therapies within the right-sided inhabitants (14.3% (95% CI, 4.8–30.3) versus 14.0% (95% CI, 5.8–26.7); OR, 1.02 (95% CI, 0.28–3.51); Prolonged Information Fig. 4). Within the general detrimental hyperselected inhabitants, the healing resection charge was larger with panitumumab (18.9% (95% CI, 14.3–24.2)) than bevacizumab (11.1% (95% CI, 7.6–15.4); OR, 1.87 (95% CI, 1.15–3.09)). In sufferers with gene alterations, the healing resection charge was practically similar with panitumumab and bevacizumab within the left-sided inhabitants (12.3% (95% CI, 5.5–22.8) versus 12.2% (95% CI, 4.6–24.8); OR, 1.01 (95% CI, 0.33–3.26)) however trended larger with panitumumab within the right-sided inhabitants (9.3% (95% CI, 2.6–22.1) versus 4.9% (95% CI, 0.6–16.5); OR, 2.00 (95% CI, 0.37–15.0); Prolonged Information Fig. 4). Within the general gene-altered inhabitants, the healing resection charge was 11.0% (95% CI, 5.8–18.4) with panitumumab and eight.5% (95% CI, 3.7–16.1) with bevacizumab (OR, 1.33 (95% CI, 0.53–3.54)).
Outcomes by RAS/BRAF and microsatellite stability standing
Present clinically adopted biomarkers (RAS/BRAF and microsatellite secure (MSS) standing) within the first-line mCRC inhabitants have been additionally explored. Amongst 733 ctDNA-evaluable sufferers, 598 sufferers (81.6%) have been WT for RAS and BRAF and have been MSS or had low microsatellite instability (MSI-L), together with 497 (67.8%) with left-sided main tumors and 96 (13.1%) with right-sided main tumors (Fig. 1 and Supplementary Desk 2). A complete of 135 sufferers (18.4%) had BRAF V600E (78 sufferers (10.6%)) and/or RAS mutations (53 sufferers (7.2%)) and/or MSI-H (20 sufferers (2.7%)).
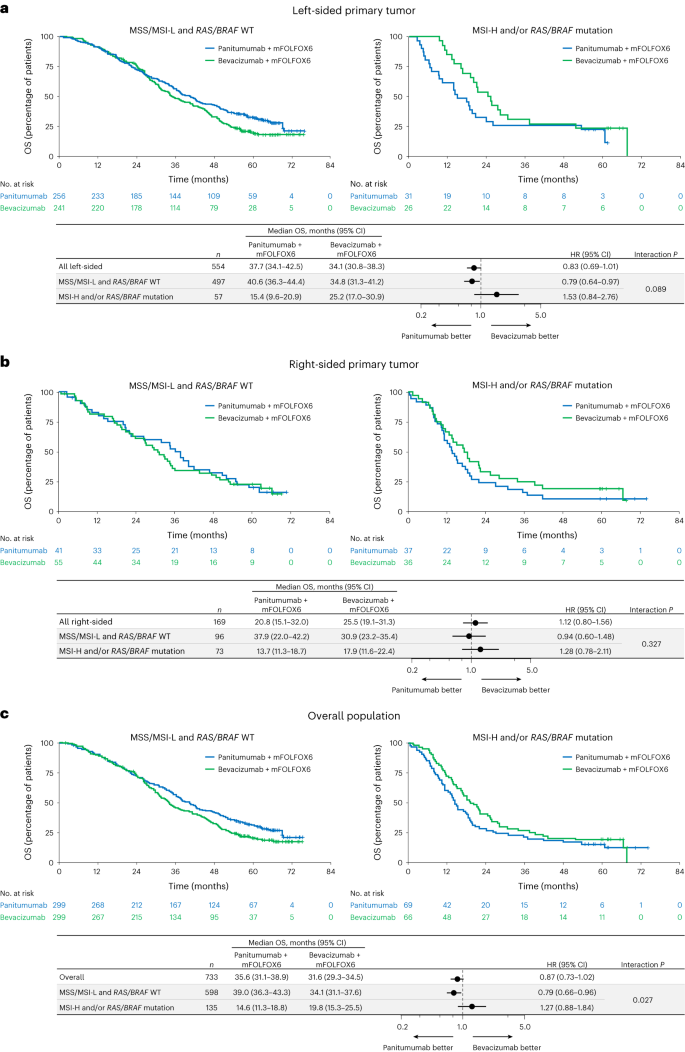
Median OS between panitumumab versus bevacizumab in sufferers with RAS/BRAF WT and MSS/MSI-L was 40.6 versus 34.8 months (HR, 0.79; 95% CI, 0.64–0.97; interplay P = 0.089; Fig. 5a) within the left-sided, 37.9 versus 30.9 months (HR, 0.94; 95% CI, 0.60–1.48; interplay P = 0.327; Fig. 5b) within the right-sided and 39.0 versus 34.1 months (HR, 0.79; 95% CI, 0.66–0.96; interplay P = 0.027; Fig. 5c) within the general populations. For sufferers with RAS/BRAF mutation or MSI-H, median OS was inferior or related with panitumumab versus bevacizumab within the left-sided (15.4 versus 25.2 months; HR, 1.53; 95% CI, 0.84–2.76; Fig. 5a), right-sided (13.7 versus 17.9 months; HR, 1.28; 95% CI, 0.78–2.11; Fig. 5b) and general populations (14.6 versus 19.8 months; HR, 1.27; 95% CI, 0.88–1.84; Fig. 5c).
a–c, Kaplan–Meier estimates of OS in sufferers with left-sided main tumors (a), sufferers with right-sided main tumors (b) and the general inhabitants (all ctDNA-evaluable sufferers) (c). The forest plots under every Kaplan–Meier plot present HR ± 95% CI. A Cox proportional hazard mannequin with out stratification elements was used to calculate HRs for group comparisons and P values for the interplay between detrimental hyperselection standing and remedy group. Statistical checks have been two-sided with out adjustment for a number of comparisons.
Median PFS was comparable between panitumumab and bevacizumab for RAS/BRAF WT and MSS/MSI-L sufferers however tended to be shorter with panitumumab than bevacizumab in sufferers with a RAS/BRAF mutation and/or MSI-H, no matter tumor sidedness (Prolonged Information Fig. 5). Antitumor response charges (Prolonged Information Fig. 6) and depth of response (Supplementary Desk 3 and Prolonged Information Fig. 7) tended to enhance with panitumumab versus bevacizumab in RAS/BRAF WT and MSS/MSI-L sufferers and poorer with panitumumab than bevacizumab for sufferers with a RAS/BRAF mutation and/or MSI-H, no matter sidedness. Response charges are proven by particular gene alteration in Supplementary Fig. 1. Healing resection charges are proven by RAS/BRAF and MSS standing in Prolonged Information Fig. 8.
Security
Antagonistic occasions occurred in 98.6% of sufferers within the biomarker inhabitants (Prolonged Information Desk 2). The incidence of hostile occasions and grade 3 or larger hostile occasions was related in detrimental hyperselected and gene-altered sufferers in every remedy group. An identical development was noticed when the triple-negative group (RAS/BRAF WT and MSS/MSI-L) was in contrast with the mutation (RAS/BRAF mutation and MSI-H) group (Supplementary Desk 4).