Allelic steadiness of mutant and wild-type KRAS impacts homoeostasis within the murine gut
To precisely mannequin the contribution of wild-type KRAS in CRC, we designed a GEMM that permits selective deletion of the wild-type Kras whereas expressing an oncogenic KrasLSL-G12D allele (hereafter KrasG12D). Right here, the wild-type Kras allele is changed by a conditional Krasflox allele to generate Krasfl/LSL-G12D (hereafter known as Krasfl/G12D) (Fig. 1a). Recombination of those alleles is focused to the intestinal epithelium by way of exercise of a tamoxifen-inducible Cre recombinase, expressed below the management of the Villin promoter (villin-creERT2) (Fig. 1a). Utilizing this mannequin we confirmed the influence of the oncogenic KrasG12D mutation upon intestinal epithelial homoeostasis, and subsequently went on to characterise any modification of this phenotype elicited by Krasfl/G12D. Intestinal tissue was sampled from mice at 30 days post-induction, with mutant KRAS discovered to advertise enterocyte proliferation and robustly suppress Paneth cell differentiation within the small gut as a consequence of elevated MAPK signalling, per earlier stories25 (Fig. 1b–e). These options are additional exacerbated by deletion of the wild-type copy in Krasfl/G12D mice, exemplified by elevated proliferation within the intestinal crypt (BrdU+) (Fig. 1d, e). Furthermore, Lysozyme and periodic acid-Schiff (PAS) or Alcian blue (AB) stains indicated additional suppression of the Paneth cell lineage and elevated abundance of secretory goblet cells (Fig. 1d, e). These in vivo knowledge are suggestive of a task for wild-type KRAS in restraining the influence of oncogenic KRAS mutation on the intestinal epithelium, which in flip interprets into quantitative phenotypic variations.
a Schematic representing the era of Krasfl/G12D mice: Villin Cre, Cre recombinase; ER oestrogen receptor; lox, Cre-Lox recombination web site; LSL Lox-Cease-Lox cassette. Created with BioRender.com. b Variety of crypts per circumference from no less than 25 of the small gut of Kras+/+, Kras+/G12D and Krasfl/G12D mice, sampled 30 days put up Cre-induction (Kras+/+, n = 3, 2M,1F; Kras+/G12D, n = 4, 2M, 2F; and Krasfl/G12D, n = 5, 4M, 1F). Knowledge are ± s.e.m. *P = 0.0286, **P = 0.0079, one-way Mann–Whitney U check. Experiments carried out on C57BL/6J background ≥ N2. c Size (cm) of small gut (SI) and colon of Kras+/+, Kras+/G12D and Krasfl/G12D mice, sampled 30 days put up Cre-induction (Kras+/+, n = 4, 3M, 1F; Kras+/G12D, n = 4, 2M, 2F; and Krasfl/G12D, n = 5, 3M, 2F). Knowledge are imply ± s.e.m. *P = 0.0143, **P = 0.0079 one-way Mann–Whitney U check. Experiments carried out on C57BL/6J background ≥ N2. d Consultant 5-bromo-2′-deoxyuridine (BrdU), Lysozyme, Alcian blue/periodic acid-Schiff (AB/PAS) and H&E staining of Kras+/+, Kras+/G12D and Krasfl/G12D mouse small gut, sampled 30 days put up Cre-induction. Scale, 100 μm. e High: Variety of BrdU-positive cells from no less than 25 half-crypts in SI from (d). Knowledge are imply ± s.e.m, (Kras+/+, n = 3, 2M, 1F; Kras+/G12D, n = 5, 3M, 2F; Krasfl/G12D n = 5, 3M, 2F), **P = 0.004, one-way Mann–Whitney U check. Center: Variety of Lysozyme-positive cells from no less than 25 crypts in SI of Kras+/+, Kras+/G12D and Krasfl/G12D in SI from (d) Knowledge are imply ± s.e.m, (Kras+/+, n = 3, 2M, 1F; Kras+/G12D, n = 6, 3M, 3F; Krasfl/G12D n = 5, 3M, 2F), **P = 0.0022, one-way Mann–Whitney U check. Backside: Variety of PAS-positive cells from no less than 25 half-crypts in SI. Kras+/+, Kras+/G12D and Krasfl/G12D in SI from (d) Knowledge are imply ± s.e.m, (Kras+/+, n = 3, 2M, 1F; Kras+/G12D, n = 6, 4M, 2F; Krasfl/G12D n = 6, 4M, 2F). **P = 0.0011, one-way Mann–Whitney U check. Experiments carried out on C57BL/6J background ≥ N2. Supply knowledge are supplied as a Supply Knowledge file.
Allelic imbalance is reported to be a typical characteristic related to many oncogenes along with KRAS1. For instance, BRAF allelic imbalance is reported to happen in 40% of BRAF mutant pores and skin cancers26. Due to this fact, we examined whether or not Braf allelic imbalance may additionally influence intestinal homoeostasis. Right here, we assessed the influence of a conditional oncogenic BrafV600E allele, alone or together with a conditional Braf focusing on allele (henceforth known as Braffl/V600E), or when bred to homozygosity (BrafV600E/V600E), once more below the management of villin-creERT2. To evaluate the influence of BrafV600E gene dosage, intestinal tissues had been sampled from Braf+/V600E, Braffl/V600E, BrafV600E/V600E or management mice at a time level 3 days post-induction recombination. As beforehand reported, we discovered that BrafV600E/+ promotes proliferation within the intestinal crypt (BrdU+), certainly to a better diploma than KrasG12D/+ presently level (Supplementary Fig. 1a, b)27,28. In step with the phenotypes noticed in KRAS mutant gut, we discover that altering allelic steadiness in favour of oncogenic Braf, both by way of breeding to homozygosity (BrafV600E/V600E), or by way of conditional deletion of the wild-type allele (Braffl/V600E) results in vital enhance in crypt cell proliferation and lack of Lysozyme-positive Paneth cells (Supplementary Fig. 1a, b). Nevertheless, BRAF activation doesn’t considerably alter the abundance of secretory goblet cells within the intestinal epithelium (Supplementary Fig. 1a, b). Collectively, these knowledge present that allelic imbalance at oncogene loci akin to that seen in human most cancers exacerbates particular oncogene-associated and cancer-related phenotypes.
Wild-type Kras deletion within the presence of oncogenic KRASG12D and Wnt activation accelerates tumorigenesis by way of MAPK signalling
Given the noticed influence upon regular intestinal homoeostasis pushed by altered gene dosage at oncogenic loci, we investigated whether or not these occasions may cooperate with the concomitant lack of the tumour suppressor gene Apc. The Krasfl/G12D mouse line described above was bred to the well-characterised mouse line bearing conditional deletion of the tumour suppressor gene Apc (villin-creERT2 Apcfl/fl), producing Apcfl/fl Krasfl/G12D or Apcfl/fl Kras+/G12D mouse strains as easy, tractable fashions of oncogene-induced hyperproliferation in vivo. Homozygous deletion of Apc within the murine gut leads to a sturdy phenotype, pushed by hyperproliferation and altered differentiation of the intestinal crypt epithelium29. Furthermore, we now have proven that this phenotype is exacerbated by expression of oncogenic Kras10,11. Utilizing this technique, we investigated whether or not lack of wild-type Kras, within the context of an oncogenic KrasG12D mutation and Apc loss, impacted epithelial proliferation in vivo. Certainly, this was the case with considerably enhanced proliferation, as denoted by BrdU incorporation, noticed within the intestinal epithelium of Apcfl/fl Krasfl/G12D mice when in comparison with Apcfl/fl Kras+/G12D mice (Fig. 2a, b), with the world of proliferative cells extending greater within the villus epithelium in Apcfl/fl Krasfl/G12D mice. This was not as a consequence of Kras copy quantity or allele modifications as confirmed utilizing droplet PCR (Supplementary Fig. 2a, b). The ectopic proliferation/dedifferentiation of cells within the villus epithelium is a key characteristic of KrasG12D mutation within the context of Apc deficiency, and is concomitant with an acquired capability of mutant cells to kind organoid cultures in vitro11. In step with the noticed proliferation within the villus epithelium of Apcfl/fl Krasfl/G12D mice, the attribute organoid forming capability was enhanced when in comparison with Apcfl/fl Kras+/G12D mice (Supplementary Fig. 2c, d). These knowledge point out that lack of the wild-type copy of Kras within the context of concomitant oncogenic KrasG12D mutation and Apc depletion can improve a variety of KrasG12D related phenotypes.
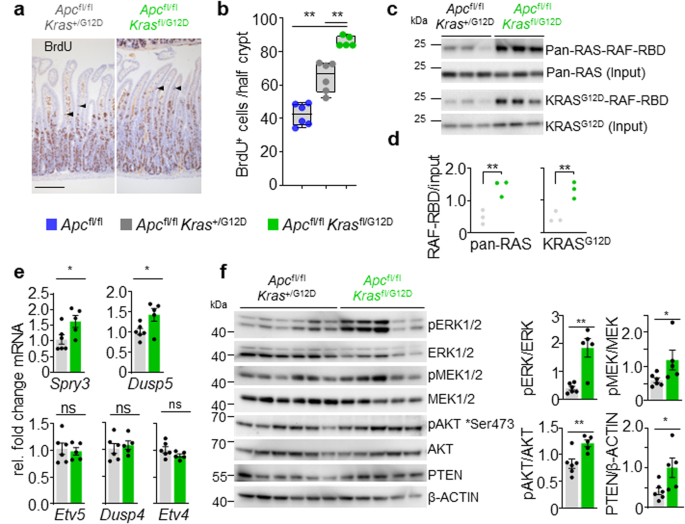
a Consultant H&E and BrdU IHC of Apcfl/fl Kras+/G12D and Apcfl/fl Krasfl/G12D mice sampled 3 days put up Cre-induction. Arrowheads point out de-differentiating cells within the villi. Scale, 100 μm. b Boxplots exhibiting BrdU-positive cells from no less than 25 half-crypts in SI in Apcfl/fl, Apcfl/fl Kras+/G12D and Apcfl/fl Krasfl/G12D mice. Bins depict interquartile vary, central line signifies median and whiskers point out minimal/most values (Apcfl/fl, n = 6, 4M, 2F; Apcfl/fl Kras+/G12D, n = 6, 2M, 4F; and Apcfl/fl Krasfl/G12D, n = 5, 1M, 4F mice). **P = 0.0022, **P = 0.0043. c RAF-Ras Binding Area (RBD) agarose affinity purification assay of three biologically impartial samples per situation from Apcfl/fl Kras+/G12D and Apcfl/fl Krasfl/G12D intestinal organoids. Pulldown of RAS-GTP with RAF-RBD agarose beads. High: Precipitates had been immunoblotted utilizing a pan-RAS antibody and enter pan-RAS served as loading management. RAS-GTP activation ranges had been quantified and normalised to pan-Ras loading management. Apcfl/fl Kras+/G12D, n = 3, 2M, 1F; and Apcfl/fl Krasfl/G12D, n = 3, 1M, 2F. Backside: Precipitates had been immunoblotted utilizing a KRASG12D antibody and enter KRASG12D served as loading management. KRASG12D-RAF-RBD ranges had been quantified and normalised to KRASG12D loading management. Apcfl/fl Kras+/G12D, n = 3, 3M; and Apcfl/fl Krasfl/G12D, n = 3, 1M, 2F. d Quantification of RAF-RBD assay (left) and KRASG12D immunoblot (proper from c), **P = 0.0045 (pan-RAS), **P = 0.0044 (KRASG12D). e qRT-PCR evaluation of Etv5, Etv4, Dusp4, Dusp5 and Spry3 in Apcfl/fl Kras+/G12D (n = 6, 3M, 3F) and Apcfl/fl Krasfl/G12D (n = 5, 1M, 4F) intestinal organoids. Knowledge are imply ± s.e.m. Transcript ranges had been normalised to Gapdh. *P = 0.0260 (Spry3) *P = 0.0411 (Dusp5). f Left: Immunoblots exhibiting PTEN, pAKT (Ser473), AKT, pERK1/2, ERK1/2, pMEK1/2, MEK1/2 and ß-actin in Apcfl/fl Kras+/G12D and Apcfl/fl Krasfl/G12D intestinal organoids. 6 Apcfl/fl Kras+/G12D and 5 Apcfl/fl Krasfl/G12D organic replicates per group, every lane represents organoids generated from particular person mice from genotype indicated. Proper: Quantification of immunoblots, phosphorylated proteins had been normalised to whole protein ranges. Knowledge are imply ± s.e.m. **P = 0.0043 (pERK/ERK), *P = 0.0152 (pMEK/MEK), **P = 0.0087 (pAKT/AKT), *P = 0.015 (PTEN/ß-actin). b, d, e, f utilizing one-way Mann–Whitney U check. Supply knowledge are supplied as a Supply Knowledge file.
We hypothesised that lack of the wild-type Kras allele could alter baseline gene dosage of the oncogenic mutant allele, and because of this, result in enhanced KRAS activation and elevated signalling flux by way of downstream effector pathways and any related suggestions loops. This speculation was examined by way of comparability of organoid cultures derived from Apcfl/fl Krasfl/G12D and Apcfl/fl Kras+/G12D intestinal tissues. Initially, we quantified the relative proportion of GTP-bound, lively RAS proteins. In these pull-down assays, Apcfl/fl Krasfl/G12D organoids had been characterised by enhanced binding of RAS to the RAS-binding area (RBD) of BRAF, when in comparison with Apcfl/fl Kras+/G12D, suggestive of a bigger pool of lively RAS (Fig. 2c, d). It’s well-known that elevated RAS exercise interprets into elevated activation of downstream effector pathways such because the MAPK cascade30. To find out whether or not MAPK exercise was elevated in Apcfl/fl Krasfl/G12D and Apcfl/fl Kras+/G12D organoids, we quantified expression of recognized ERK-regulated transcripts, discovering these to be enriched in Apcfl/fl Krasfl/G12D organoids (Fig. 2e). To judge MAPK exercise in additional depth, we assessed downstream effector exercise in these strains by way of quantitative immunoblotting. We detected a transparent enhance in phosphorylation of ERK1/2 and MEK1/2 in Apcfl/fl Krasfl/G12D organoids, with phosphorylation of AKT and expression of PTEN additionally elevated (Fig. 2f). These knowledge point out that lack of the wild-type copy of Kras enhances the relative exercise of the oncogenic mutant allele and drives downstream effector pathway signalling.
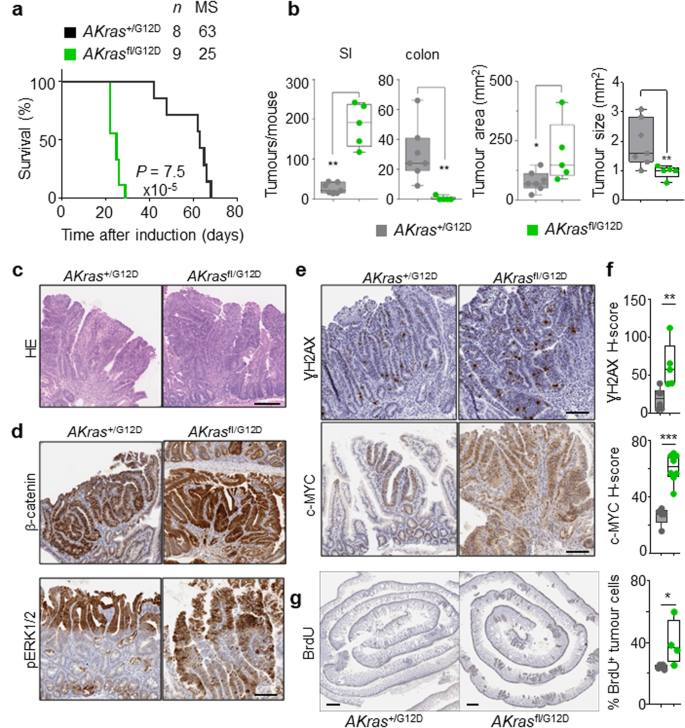
Given the strong influence of wild-type Kras deletion within the acute setting above, we subsequent addressed the position of wild-type KRAS on oncogenic KRASG12D-driven intestinal tumorigenesis. To this finish, we generated villin-creERT2 Apcfl/+ Kras+/G12D (henceforth AKras+/G12D) and villin-creERT2 Apcfl/+ Krasfl/G12D (henceforth AKrasfl/G12D). On this setting, intestinal tumour improvement happens following sporadic lack of the second copy of Apc in particular person intestinal crypts. Focused mutation within the intestinal epithelium was induced by way of intraperitoneal administration of tamoxifen, with deletion of wild-type Kras within the context of oncogenic KrasG12D (AKrasfl/G12D) leading to a major acceleration of tumorigenesis, and a consequent discount of median survival based mostly upon an endpoint outlined by medical indicators related to tumour burden. (Fig. 3a). The lowered time to onset of indicators related to intestinal tumorigenesis in AKrasfl/G12D mice was coincident with a hanging tumour initiation phenotype, with improvement of quite a few small lesions noticed principally within the small gut (Fig. 3b). The initiating lesions noticed in AKrasfl/G12D mice had been histopathologically corresponding to these noticed in AKras+/G12D mice (Fig. 3c), and as anticipated, had been optimistic for nuclear β-catenin (Fig. 3d). Immunohistochemical evaluation demonstrated nuclear accumulation of phosphorylated ERK1/2 (Fig. 3d), elevated expression of c-MYC and elevated abundance of γH2AX in AKrasfl/G12D tumours when in comparison with AKras+/G12D tumours, suggestive of MAPK pathway activation, elevated mobile proliferation and activation of DNA injury response pathways (Fig. 3e, f). Certainly, utilizing BrdU incorporation as a marker for mobile proliferation, we demonstrated that the tumour epithelium of AKrasfl/G12D mice is markedly extra proliferative than that of AKras+/G12D mice (Fig. 3g). Collectively, these outcomes present that lack of the wild-type copy of Kras will increase exercise and signalling of the oncogenic mutant allele driving tumour initiation.
a Kaplan–Meier survival curve of villin-creERT2 Apc+/fl Kras+/G12D (AKras+/G12D) and villin-creERT2 Apc+/fl Krasfl/G12D (AKrasfl/G12D) mice aged till medical endpoint (Apc+/fl Kras+/G12D, n = 8, 5M, 3F, MS, median survival = 63; Apc+/fl Krasfl/G12D, n = 9, 5M, 4F, MS, median survival = 25), ****P = 7.5 × 10-5, log-rank (Mantel-Cox) check. b Left: Boxplots exhibiting whole variety of tumours from Apc+/fl Kras+/G12D and Apc+/fl Krasfl/G12D mice aged till medical endpoint in SI and Colon. Proper: Boxplots exhibiting tumour space (mm2) and tumour measurement (mm2) in Apc+/fl Kras+/G12D and Apc+/fl Krasfl/G12D mice aged till medical endpoint. Bins depict interquartile vary, central line signifies median and whiskers point out minimal/most values (Apc+/fl Kras+/G12D, n = 7, 5M, 2F; Apc+/fl Krasfl/G12D n = 5, 3M, 2F). **P = 0.0013 (SI), **P = 0.0013 (colon), *P = 0.036 (space), **P = 0.0088 (measurement), one-way Mann–Whitney U check. c Consultant H&E photos of AKras+/G12D (n = 5, 2M, 3F) and AKrasfl/G12D (n = 8, 5M, 3F) tumour. Dashed field highlights chosen space proven in excessive magnification. d Consultant photos of nuclear β-catenin and pERK1/2 staining in Apc+/fl Kras+/G12D (n = 5, 2M, 3F) and Apc+/fl Krasfl/G12D (n = 8, 5M, 3F) tumours at medical endpoint. Scale, 100 µm. e Consultant photos of γH2AX and c-MYC staining in Apc+/fl Kras+/G12D (n = 5, 2M, 3F) and Apc+/fl Krasfl/G12D (n = 8, 5M, 3F) tumours at medical endpoint. Scale, 100 µm. f H-score of γH2AX (Apc+/fl Kras+/G12D, n = 6, 4M, 2F; Apc+/fl Krasfl/G12D n = 5, 3M, 2F) and c-MYC (Apc+/fl Kras+/G12D, n = 5, 2M, 3F; Apc+/fl Krasfl/G12D n = 8, 5M, 3F) IHC staining of (e). Bins depict interquartile vary, central line signifies median and whiskers point out minimal/most values. **P = 0.0043, ***P = 0.0008, one-way Mann–Whitney U check. g Consultant BrdU staining of Apc+/fl Kras+/G12D and Apc+/fl Krasfl/G12D small intestinal tumours at medical endpoint. Scale, 100 µm. Proper: quantification of BrdU positivity in tumour cells. Bins depict interquartile vary, central line signifies median and whiskers point out minimal/most values. (Apc+/fl Kras+/G12D, n = 5, 3M, 2F; Apc+/fl Krasfl/G12D n = 4, 2M, 2F). *P = 0.0317, one-way Mann–Whitney U check. Supply knowledge are supplied as a Supply Knowledge file.
Lack of wild-type KRAS restores sensitivity to MEK inhibition in colorectal tumours in vivo
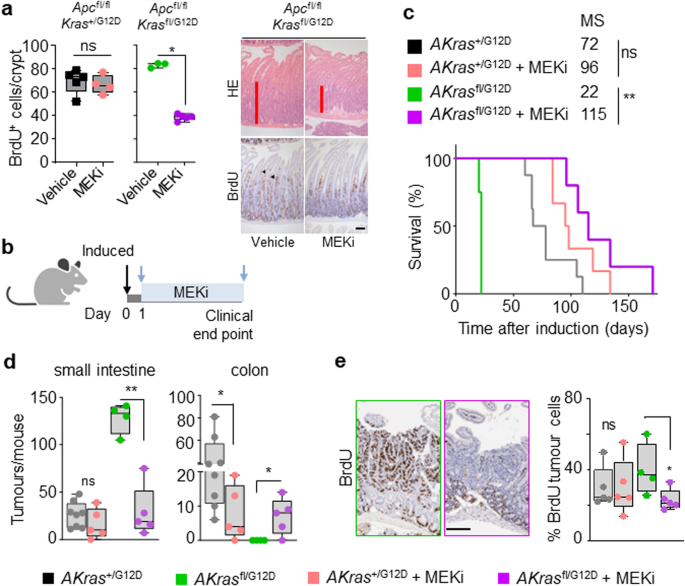
We’ve demonstrated that altered allelic steadiness of oncogenic KrasG12D has a considerable influence upon tumour initiation in fashions of intestinal illness. On condition that KRAS mutation is clinically related to resistance to focused therapies, we subsequent investigated whether or not therapeutic efficacy is positively or negatively influenced by allelic imbalance on the Kras locus. We’ve beforehand proven that the noticed intestinal crypt epithelium hyperproliferation attribute of the Apcfl/fl Kras+/G12D mannequin is immune to MEK inhibition11,31, and that this proliferation is enhanced in Apcfl/fl Krasfl/G12D mice concomitant with elevated KRAS and MAPK exercise (Fig. 2c, f). We reasoned that this elevated MAPK activation may end in an acquired sensitivity to inhibition of MEK1/2 with a clinically related focused therapeutic agent (AZD6244/selumetinib). As beforehand, we discovered that remedy of Apcfl/fl Kras+/G12D mice with AZD6244 had no influence upon intestinal hyperproliferation. Importantly, and in distinction, remedy with AZD6244 not solely considerably decreased proliferation within the intestinal crypt epithelium of Apcfl/fl Krasfl/G12D mice however suppressed proliferation to a degree under that of vehicle-treated Apcfl/fl Kras+/G12D mice (Fig. 4a). We subsequent examined whether or not MEK1/2 inhibition had the same suppressive impact upon the method of intestinal tumourigenesis. To do that, we handled AKrasfl/G12D or AKras+/G12D mice with AZD6244 (25mgkg-1, BID) from 1-day post-induction of genetic recombination (Fig. 4b). We discovered that inhibition of MEK1/2 had a modest influence on the survival of AKras+/G12D mice, based mostly upon endpoint outlined by onset of medical indicators related to tumour burden, however considerably prolonged survival in AKrasfl/G12D mice (median survival prolonged from 26 days to 115 days) (Fig. 4c). This extension in survival was accompanied by a dramatic discount within the variety of small intestinal tumours (Fig. 4d) alongside enhance in colonic tumour quantity, albeit at vastly elevated time post-induction. A major discount in tumour cell proliferation (BrdU incorporation) was additionally noticed in AKrasfl/G12D mice in comparison with AKras+/G12D derived tumours (Fig. 4e). These knowledge counsel that wild-type Kras acts to suppress the penetrance of mutant oncogenic KrasG12D, thus dampening MAPK signalling and contributing to therapeutic resistance in Kras mutant tumours, a key medical downside.
a Left, quantification of BrdU-positive cells per half crypt in Apcfl/fl Kras+/G12D and Apcfl/fl Krasfl/G12D mice 3 days post-induction handled with Car or MEKi (AZD6244) as indicated (Apcfl/fl Kras+/G12D Car, n = 5, 1M, 4F; MEKi, n = 4, 1M, 3F; and Apcfl/fl Krasfl/G12D Car, n = 3, 1M, 2F; MEKi, n = 5, 3M, 2F). *P = 0.017. Proper, consultant H&E and BrdU photos of Apcfl/fl Krasfl/G12D 3 days post-induction handled with Car or MEKi (AZD6244) as indicated. Arrowheads present BrdU+ve hyper proliferative cells. Experiments carried out on C57BL/6J background ≥ N2. b Schematic presenting experimental strategy. Apc+/fl Kras+/G12D and Apc+/fl Krasfl/G12D mice handled with MEKi sooner or later post-induction and handled to medical endpoint. Created with BioRender.com. c Kaplan–Meier survival curve of Apc+/fl Kras+/G12D and Apc+/fl Krasfl/G12D mice handled as proven in (b) aged till medical endpoint (Apc+/fl Kras+/G12D, n = 8, 2M, 6F, MS, median survival = 72; Apc+/fl Kras+/G12D MEKi, n = 6, 2M, 4F, MS, median survival = 96; Apc+/fl Krasfl/G12D, n = 4, 2M, 2F, MS, median survival = 22; Apc+/fl Krasfl/G12D MEKi, n = 5, 4M, 1F, MS, median survival = 115). **P = 0.0050, ns not vital, log-rank (Mantel-Cox) check. d Boxplots exhibiting whole variety of tumours from Apc+/fl Kras+/G12D and Apc+/fl Krasfl/G12D mice untreated or handled with MEKi as indicated in (c) and aged till medical endpoint in SI and Colon (Apc+/fl Kras+/G12D, n = 8, 2M, 6F; Apc+/fl Kras+/G12D MEKi, n = 5, 2M, 3F; Apc+/fl Krasfl/G12D, n = 4, 2M, 2F; Apc+/fl Krasfl/G12D MEKi, n = 5, 4M, 1F). **P = 0.0079 (Apc+/fl Krasfl/G12D SI), *P = 0.0281, *P = 0.0397 (colon). e Left: Consultant BrdU photos of Apc+/fl Kras+/G12D and Apc+/fl Krasfl/G12D tumours from mice handled with MEKi from day 1 post-induction till medical endpoint. Proper: boxplot exhibiting proportion of BrdU-positive tumour cells in Apc+/fl Kras+/G12D and Apc+/fl Krasfl/G12D handled with MEKi (Apc+/fl Kras+/G12D automobile, n = 5, 1M, 2F; Apc+/fl Kras+/G12D MEKi, n = 5, 2M, 3F; Apc+/fl Krasfl/G12D untreated, n = 4, 2M, 2F; Apc+/fl Krasfl/G12D MEKi, n = 5, 4M, 1F), *P = 0.0159. a, d and e Bins depict interquartile vary, central line signifies median and whiskers point out minimal/most values, one-way Mann–Whitney U check. Scale, 100 μm. Supply knowledge are supplied as a Supply Knowledge file.
Lack of wild-type KRAS potentiates oncogenic KRASG12D pushed tumorigenesis within the absence of exogenous WNT mutations in vivo
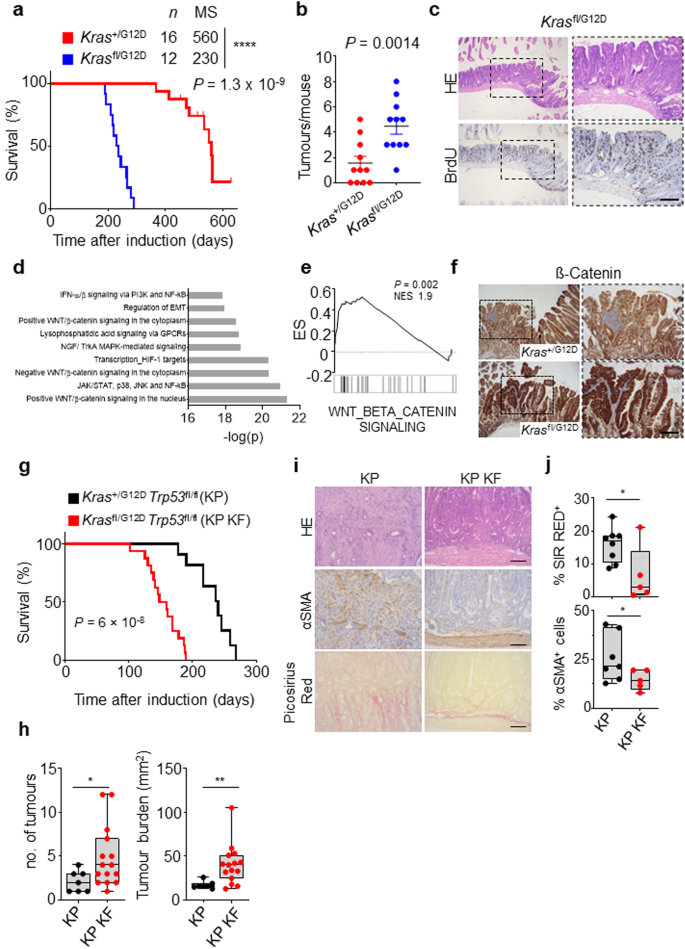
Inside the intestinal epithelium KRAS mutation alone just isn’t enough to drive tumourigenesis. Certainly, in human, KRAS mutant clones are recognized to generally come up with age, within the intestinal epithelium in morphologically regular crypts. That is recapitulated by murine fashions the place expression of an oncogenic KrasG12D mutant allele in isolation all through the intestinal epithelium (villin-creERT2 Kras+/G12D) leads to lowly penetrant intestinal lesion improvement, which interprets to an general survival of better than 12 months. Given the influence of mixed deletion of wild-type Kras and expression of oncogenic KrasG12D on each intestinal homoeostasis, and tumour initiation within the context of heterozygous lack of Apc, we subsequent requested whether or not these alterations in Kras allelic steadiness may drive tumour initiation in isolation. Right here, we induced genetic recombination in villin-creERT2 Krasfl/G12D or villin-creERT2 Kras+/G12D mice, and assessed survival based mostly upon sampling as a consequence of onset medical indicators related to vital intestinal tumour burden. As anticipated, villin-creERT2 Kras+/G12D alone resulted in inefficient tumour initiation, with median survival for this group at round 16 months. Strikingly, mice within the villin-creERT2 Krasfl/G12D group developed tumours far more quickly than the management group, exhibiting a vastly lowered median survival of round 230 days/9 months (Fig. 5a). The tumours from each Krasfl/G12D and Kras+/G12D mice had been predominantly discovered within the small gut, and had been characteristically giant adenomas. Nevertheless, tumour quantity was considerably elevated within the small gut of Krasfl/G12D mice (Fig. 5b, c). This demonstrates that even when the one exogenously launched driver mutation is KrasG12D, lack of wild-type Kras considerably promotes intestinal tumourigenesis, which in flip interprets into considerably shortened survival.
a Kaplan–Meier survival curve of villin-creERT2 Kras+/G12D and Krasfl/G12D mice aged till medical endpoint (Kras+/G12D, n = 16, 5M, 11F; Krasfl/G12D, n = 12, 4M, 8F). ****P = 1.3 × 10-9. MS, median survival. b Dotplot exhibiting whole variety of tumours from Kras+/G12D and Krasfl/G12D 2D mice. Knowledge are imply ± s.e.m. (Kras+/G12D, n = 11, 3M, 8F; Krasfl/G12D, n = 11, 4M, 7F). **P = 0.0014. c Consultant H&E and BrdU IHC photos of Krasfl/G12D mice. Consultant of 6 organic replicates per genotype. Dashed containers spotlight chosen areas proven in excessive magnification. Scale, 200 µm. d Metacore Community evaluation of differentially expressed genes from Krasfl/G12D tumours. e Geneset enrichment plot for Wnt beta-catenin signalling signature from the ‘Hallmark’ geneset assortment of tumours derived from Krasfl/G12D mice. X-axis reveals normalised enrichment rating (NES), and the P worth (computed and corrected for a number of testing utilizing the Benjamini–Hochberg process). f Consultant β-catenin IHC staining in Kras+/G12D and Krasfl/G12D mice. Consultant of 5 organic replicates per genotype. Dashed containers spotlight chosen areas proven in excessive magnification. Scale, 200 µm. g Kaplan–Meier survival curve of Kras+/G12D Trp53fl/fl (KP) and Krasfl/G12D Trp53fl/fl (KP KF) mice aged till medical endpoint (KP, n = 10, 7M, 3F; KP KF, n = 16, 6M, 10F) ****P = 6 × 10-8. h Boxplots exhibiting whole variety of tumours (left) and tumour burden, mm2 (proper) in KP and KP KF mice (KP, n = 7, 2M, 5F, KP KF, n = 15, 6M, 9F). *P = 0.02, **P = 0.0017. Please be aware management KP cohort utilized in (g) and (h) comprise completely different mice. i Consultant H&E, αSMA (alpha-smooth muscle actin) and Sirius Pink staining of KP and KP KF tumours. Consultant of KP n = 8; KP KF n = 5. Scale, 100 µm. j Boxplots exhibiting Sirius Pink positivity (%) and αSMA optimistic cells (%) of KP and KP KF tumours (KP n = 7; KP KF, n = 5). *P = 0.0326 (Sirius Pink), *P = 0.024 (αSMA). h and j containers depict interquartile vary, central line signifies median and whiskers point out minimal/most values. P values in a and g are log-rank (Mantel-Cox) checks. b, h and j are one-way Mann–Whitney U checks. Supply knowledge are supplied as a Supply Knowledge file.
To raised perceive the mechanistic influence of deletion of wild-type Kras within the context of oncogenic KrasG12D, we transcriptionally profiled tumours arising in villin-creERT2 Krasfl/G12D mice and in comparison with adjoining non-transformed tissue. Geneset enrichment evaluation (GSEA), recognized key oncogenic programmes related to KRAS signalling, MEK and AKT had been enriched in villin-creERT2 Krasfl/G12D tumours (Supplementary Fig. 3a). Importantly, GSEA and Metacore evaluation of differentially expressed genes confirmed an overrepresentation of WNT and β-catenin signalling pathways (Fig. 5d, e). On condition that aberrant activation of the Wnt/β-catenin pathway, and its downstream transcriptional networks, is a typical initiating occasion in CRC, and outcomes from stabilisation and nuclear accumulation of the transcriptional co-activator β-catenin, we interrogated tumours arising in villin-creERT2 Krasfl/G12D and villin-creERT2 Kras+/G12D mice for nuclear accumulation of β-catenin. In step with the transcriptional enrichment of Wnt signalling pathways (Fig. 5e), tumours arising in Krasfl/G12D intestines confirmed positivity for nuclear β-catenin whereas Kras+/G12D intestines extra predominantly exhibited membranous β-catenin (Fig. 5f).
Lack of wild-type KRAS alters tumour development and metastasis of aggressive mutant KRAS-driven colorectal tumours
To date, we now have solely investigated the position of wild-type KRAS in mouse fashions of adenoma. Latest stories have proven that modifications in KRAS gene dosage alter the clonal evolution of tumours and alter the metastatic incidence in KRAS mutant cancers23. Given the dramatic position we now have noticed in tumour initiation, we subsequent wished to see how necessary wild-type KRAS could be in tumour development. We’ve beforehand demonstrated that deletion or mutation of the tumour suppressor gene Trp53 alongside activating mutation of Kras may give rise to aggressive, late-stage adenocarcinoma improvement within the gut32. Certainly, the human paralogue, TP53 is inactivated in additional than half of human colorectal cancers (TCGA). These fashions subsequently symbolize a perfect system for interrogating the influence of wild-type Kras deletion in a extra complicated, aggressive, clinically related oncogenic KRASG12D pushed illness.
To attain this, we interbred the villin-creERT2 Krasfl/G12D mouse line with a Trp53 conditional knockout allele to generate villin-creERT2 Krasfl/G12D Trp53fl/fl (henceforth known as KP KF) and villin-creERT2 Kras+/G12D Trp53fl/fl (henceforth known as KP) mice. Whereas KP mice developed on common one intestinal tumour and exhibited a median general survival of 240 days, with very lowly penetrant metastasis (Fig. 5g, h), KP KF mice developed on common 4 intestinal tumours and exhibited a median general survival of 150 days (Fig. 5g, h). Histological evaluation of tumours arising in KP KF mice steered a morphology akin to human tubulovillous adenoma, in distinction to the tumours which arose in management KP mice, which exhibit a serrated morphology. Furthermore, proof of native invasion or poor differentiation, options usually related to superior illness, had been obvious in KP tumours however absent from KP KF tumours (Fig. 5i). As well as, there have been clear variations within the stromal microenvironment of the KP KF tumours, corresponding to an absence of infiltrating alpha-smooth muscle actin (αSMA)-positive stromal cells and low ranges of stromal collagen deposition (as indicated by picrosirius crimson), once more, contrasted by KP tumours (Fig. 5i, j). These knowledge counsel that whereas lack of the wild-type copy of Kras in an aggressive Kras mutant-driven mannequin of intestinal most cancers can drive accelerated tumour initiation, it doesn’t endow tumours with elevated invasion or aggression, certainly these options seem suppressed.
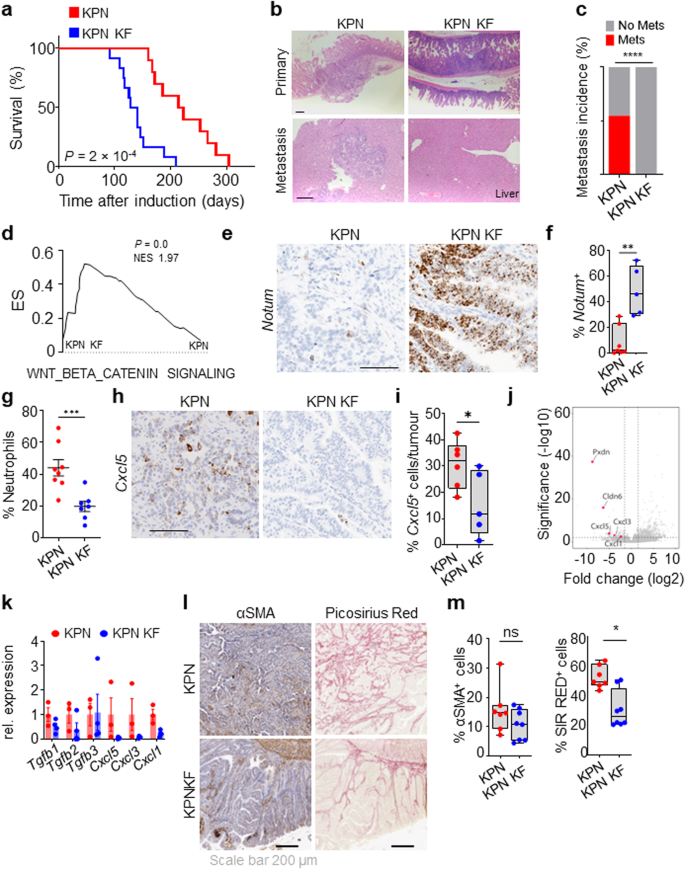
Contemplating this, we determined to additional examine the position of wild-type Kras in aggressive, late-stage KRAS-mutant CRC utilizing our not too long ago described metastatic KPN mannequin (villin-creERT2 Kras+/G12D Trp53fl/fl Rosa26N1icd/+). These KPN mice develop intestinal adenocarcinoma and exhibit extremely penetrant metastasis, predominantly to the liver32. We interbred KPN mice with KP KF mice to generate (villin-creERT2 Krasfl/G12D Trp53fl/fl Rosa26N1icd/+; henceforth known as KPN KF). Strikingly, tumorigenesis was considerably accelerated in KPN KF mice with deletion of wild-type Kras, mirrored by a major discount in general survival based mostly upon endpoint outlined by medical indicators related to tumour burden (Fig. 6a). Utilizing droplet PCR we confirmed this acceleration in tumorigenesis of KPN KF mice was not as a consequence of mutant Kras induced modifications in Kras copy quantity (Supplementary Fig. 4a, b). Comparative histological evaluation of tumours from KPN and KPN KF mice confirmed that tumours arising from KPN KF confirmed lack of native invasion with outstanding lack of metastasis incidence (Fig. 6b, c). To achieve a greater mechanistic perception into the influence of Kras deletion on this setting, we first carried out transcriptional evaluation of tumours derived from KPN KF and KPN mice. Apparently, KPN KF tumours confirmed a major enrichment of Wnt signalling pathway (Fig. 6d) and expression of the surrogate Wnt marker Notum, in comparison with KPN tumours (Fig. 6e, f). It’s notable that comparatively low ranges of Wnt activation is a key characteristic of the extremely metastatic KPN tumours, whereas comparable high-Wnt APC-deficient, APN tumours are non-metastatic32. In step with the high-Wnt activation signature in KPN KF tumours, comparative evaluation of a number of printed Wnt transcriptional signatures in tumours from APN, KPN and KPN KF mice clearly confirmed greater Wnt activation in KPN KF tumours (Supplementary Fig. 4c). Collectively, these knowledge present that Kras mutant tumours missing wild-type Kras in a KPN setting activate extra strong ranges of Wnt signalling throughout tumorigenesis. This stated, it’s noteworthy that major tumours arising within the gut of KPN KF mice are adenomatous, whereas these arising in KPN mice are of a extra aggressive adenocarcinoma/carcinomatous morphology. Whereas the underlying genetics will undoubtedly play a key position in defining the molecular make-up of the illness, the lowered histopathological grade of tumours present in KPN KF could straight affect immune and inflammatory infiltrate.
a Kaplan–Meier survival curve of Kras+/G12D Trp53fl/fl Rosa26N1icd/+ (KPN) and Krasfl/G12D Trp53fl/fl Rosa26N1icd/+ (KPN KF) mice aged till medical endpoint (KPN, n = 10, 5M, 5F, KPN KF, n = 12, 6M, 6F) ***P = 2 × 10-4, log-rank (Mantel-Cox) check. b Consultant H&E photos of major tumour and metastasis (liver) of mice from (a). Scale, 200 μm. c Incidence of metastasis (%) in KPN and KPN KF mice aged till medical endpoint (KPN, n = 11, 6M, 5F; KPN KF, n = 11, 5M, 6F) ****P = 1 × 10-15, two-tailed chi-square check. d GSEA of hallmark WNT-β-Catenin signalling in KPN KF tumours in comparison with KPN. X-axis reveals normalised enrichment rating (NES), and the P worth (computed and corrected for a number of testing utilizing the Benjamini–Hochberg process). e Consultant Notum in situ hybridisation (ISH) in KPN and KPN KF tumours. Scale, 100 μm. f Boxplots exhibiting Notum (ISH) optimistic cells per tumour (%) of KPN and KPN KF tumours (KPN, n = 7, 2M, 5F; KPN KF, n = 5, 2M, 3F). **P = 0.0013. g Dotplot exhibiting neutrophils proportion in systemic blood of KPN and KPN KF mice aged to medical endpoint (KPN, n = 8, 6M, 2F; KPN KF, n = 7, 5M, 2F). ***P = 0.0006. h Consultant Cxcl5 ISH in KPN and KPN KF tumours. Consultant of 5 mice per genotype. Scale, 100 μm. i Boxplots exhibiting Cxcl5 optimistic cells per tumour (%) of KPN and KPN KF tumours (KPN, n = 6, 2M, 4F; KPN KF, n = 5, 2M, 3F). *P = 0.0411. j Volcano plot of differentially expressed genes in organoids derived from KPN KF tumours. okay Relative expression of Tgfβ ligands and chemokines from organoids derived from KPN and KPN KF tumours. Knowledge are imply ± s.e.m. KPN, n = 3, 3M; KPN KF, n = 4, 1M, 3F. l Consultant αSMA (alpha-smooth muscle actin) and Sirius Pink staining of KPN and KPN KF tumours. Scale, 200 µm. m Boxplots exhibiting Sirius Pink positivity (%) and αSMA optimistic cells (%) of ageing or vehicle-treated KPN and KPN KF tumours (KPN n = 7, 2M, 5F; KPN KF n = 8, 4M, 4F). *P = 0.0103. f, i and m containers depict interquartile vary, central line signifies median and whiskers point out minimal/most values. f, g, i and m are one-way Mann–Whitney U checks. Supply knowledge are supplied as a Supply Knowledge file.
Nonetheless, we now have beforehand proven {that a} essential characteristic of metastasis within the Notch1-driven KPN tumours is TGFβ pathway-mediated neutrophil infiltration32. Nevertheless, we detected an absence of systemic circulating neutrophil accumulation within the blood of KPN KF mice (Fig. 6g). On condition that chemokines corresponding to Cxcl5 are implicated in neutrophil attraction, we assessed expression of Cxcl5 and located considerably lowered expression within the epithelium of KPN KF tumours (Fig. 6h, i). To find out how lack of wild-type Kras alters the TGFβ pathway and metastasis, we examined the transcriptome of tumour-derived KPN and KPN KF organoids. Apparently, we discovered that NOTCH1-mediated gene expression of Tgfb2 and chemokines, corresponding to Cxcl1, Cxcl3 and Cxcl5, was downregulated in KPN KF organoids (Fig. 6j, okay). Furthermore, GSEA evaluation confirmed lack of TGFβ signalling in KPN KF organoids (Supplementary Fig. 4d). Moreover, together with the low expression of neutrophil markers (Fig. 6h, i), Tgfb2 expression was lowered in KPN KF tumours (Supplementary Fig. 4e). As well as, we noticed clear variations within the stromal microenvironment of the KPN KF tumours, with considerably low stromal collagen deposition (picrosirius crimson), once more, contrasted by collagen excessive KPN tumours (Fig. 6l, m).
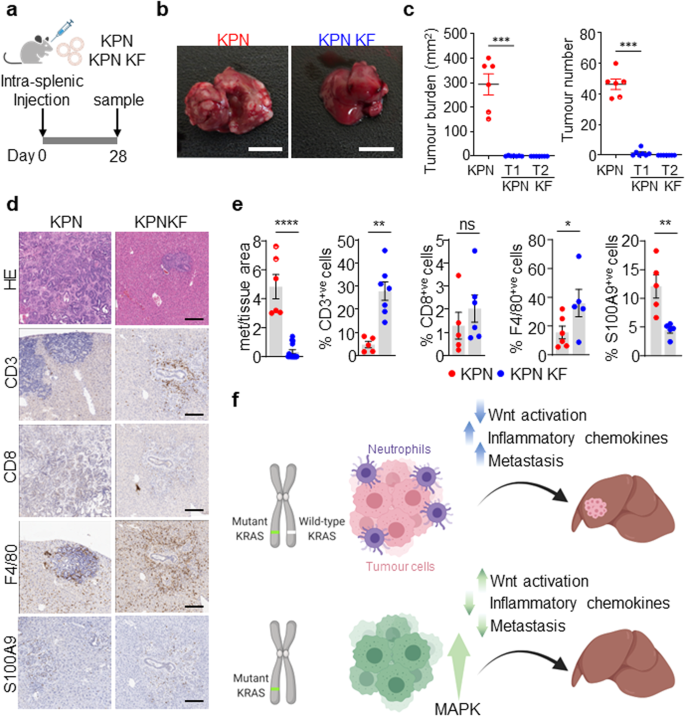
On condition that we now have beforehand proven that KPN tumour cells generate an immunosuppressive, pro-metastatic surroundings, we assessed the power of KPN KF tumour cells to supply such an immunosuppressive surroundings to generate distant metastasis. To deal with this experimentally, we carried out orthotopic intrasplenic transplantation of KPN and KPN KF tumour-derived organoids in syngeneic recipient mice (Fig. 7a). All mice with KPN organoid transplantation confirmed metastasis to both liver or lung (Fig. 7b). Nevertheless, the metastasis burden was considerably blunted in KPN KF organoid transplanted mice (Fig. 7b, c). Remarkably, we noticed a major enhance in immune cell infiltration in KPN KF transplants in comparison with the KPN management tumours (Fig. 7d, e). The noticed influence upon immune and inflammatory infiltrate was impartial of the dimensions of metastatic deposits discovered within the liver – the decreased neutrophil and elevated lymphocyte affiliation with metastatic deposits in KPN KF transplants versus KPN transplants was maintained when evaluation was restricted to tumours of comparable measurement (Supplementary Fig. 5a, b). Furthermore, the obvious influence upon immune cell infiltration was additionally confirmed within the KPN KF GEMM tumours (Supplementary Fig. 4g). Collectively, this means that lack of wild-type Kras with epithelial NOTCH1 activation in KPN tumours prompts Wnt, restricts the TGFβ mediated neutrophil recruitment required to generate an immunosuppressive, pro-metastatic area of interest and blunts the metastases of tumours missing wild-type Kras (Fig. 7f).
a Schematic exhibiting the intrasplenic transplantation of KPN and KPN KF organoids. b Photos of liver tumour burden from KPN and KPN KF organoid transplant mice. Consultant of six mice per organoid transplant. Scale, 1 cm. c Quantifications of liver tumour burden and tumour quantity from KPN and two KPN KF (T1 and T2) organoid transplants. (KPN n = 6, 6M, KPN KF T1 n = 7, 7F, KPN KF T2 n = 7, 7F). Knowledge are imply ± s.e.m. Semi-circles embrace lung metastasis burden. ***P = 0.0006, one-way Mann–Whitney U check. Created with BioRender.com. d Consultant HE, CD3, CD8, F4/80 and S100A9 IHC in KPN and KPN KF transplant mice. Consultant of six mice per organoid line. Scale, 200 μm. e Quantifications from d KPN KF T1 and T2 with tumour burden represented collectively as KPN KF. Knowledge are imply ± s.e.m. Consultant of KPN n = 6, 6M, KPN KF n = 14, 14F, ****P = 2.5 × 10-5 (Met space); KPN n = 5, 5M, KPN KF n = 7, 7F, **P = 0.0013 (CD3); KPN n = 5, 5M, KPN KF n = 6, 6F, ns, P = 0.2143 (CD8); KPN n = 6, 6M, KPN KF n = 5, 5F, *P = 0.02 (F4/80); KPN n = 5, 5M, KPN KF n = 5, 5F, **P = 0.0079 (S100A9), one-way Mann–Whitney U check. f Schematic mannequin exhibiting the position of wild-type KRAS in KRAS mutant CRC. Lack of wild-type Kras in KPN KF tumours promotes tumour initiation with WNT activation, an enhanced immune infiltrate and blunted metastasis. Created with BioRender.com. Supply knowledge are supplied as a Supply Knowledge file.