A medical trial (ChiCTR2000038995) of neoadjuvant stereotactic radiosurgery earlier than surgical resection of enormous BMs was registered to guage the security and effectiveness of preoperative GKRS remedy. The tissues of BMs of LUAD sufferers have been obtained throughout this trial.
BMs that obtained GKRS remedy have been labeled because the post-GKRS group, and people who didn’t obtain GKRS remedy have been labeled because the no-GKRS group. In response to the tissue dimension, burn diploma and the variety of blood cells, 5 post-GKRS and 5 no-GKRS tissues have been chosen for proteomics detection. There was no important distinction between the 2 teams of sufferers by way of intercourse, age, mutation, focused remedy, or chemotherapy (Desk 1). Earlier than the enrollment on this trial, no affected person had obtained radiotherapy. The 5 sufferers of post-GKRS group underwent GKRS inside median interval of 18 h (16–22 h). 4 sufferers underwent a median preoperative GKRS dose of 15.75 Gy (15–18 Gy) in 1 fraction, with the opposite affected person (G1006) receiving 24 Gy in 3 fractions.
MR and HE staining
The MR photographs and HE staining photographs of the tissue samples are proven in Fig. 1. The post-GKRS group samples have been labelled G1005, G1006, G1007, G1009, and G1010. The no-GKRS group samples have been labelled G3006, G3007, G3008, G3010, and G3019. Blended alerts, which indicated malignant neoplasms, have been proven by gadolinium-enhanced MR for all sufferers. Low sign depth across the BMs, which indicated edema, appeared in all instances to various levels. HE staining indicated that these tissues have been metastatic adenocarcinomas.
MR photographs and HE staining photographs. The post-γ group consisted of G1005, G1006, G1007, G1009, and G1010. The no-γ group consisted of G3006, G3007, G3008, G3010, and G3019. The MR photographs with the biggest neoplastic diameter have been chosen. HE staining photographs are additionally proven beneath the MR photographs for each case.
Proteins upregulated by GKRS remedy have been enriched in ribosomal proteins
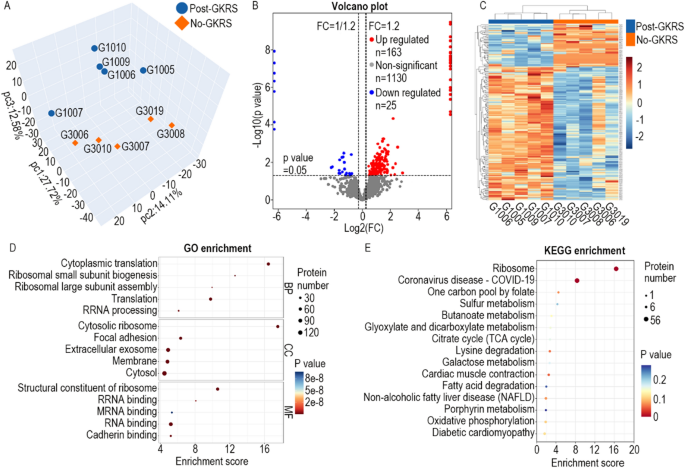
Proteomics evaluation revealed 1833 credible proteins. Principal part evaluation (PCA) was carried out primarily based on the expression of credible proteins to point out the connection amongst samples in numerous dimensions (Fig. 2A). If the distinction between two samples is important, the 2 coordinate factors are comparatively distant on the rating chart. Though the samples in every group differed from one another, the ten samples have been roughly labeled into two teams in response to their GKRS historical past. Distinction evaluation revealed that 25 proteins have been downregulated and 163 proteins have been upregulated by GKRS therapy (Fig. 2B). Cluster evaluation confirmed that the clusters of those differentially expressed proteins (DEPs) have been per the PCA outcomes (Fig. 2C).
Proteins upregulated by gamma-knife remedy have been enriched in ribosomal proteins. (A) PCA rating chart. Every level within the chart represents a pattern. The space between completely different samples will be seen visually, and the similarity or distinction within the samples will be judged accordingly. (B) The volcano plot exhibits 25 downregulated proteins (blue) and 163 upregulated proteins (crimson). (C) Heatmap displaying that the DEPs clustered into the right teams. (D) Prime 5 organic course of (BP), mobile part (CC), and molecular operate (MF) phrases. (E) Prime 15 KEGG pathway enrichment phrases. FC, fold change.
GO enrichment and KEGG pathway enrichment have been carried out to discover the doable features of those DEPs. Primarily based on organic course of (BP) evaluation, these DEPs have been enriched in cytoplasmic translation, ribosomal small subunit biogenesis, ribosomal massive subunit meeting, translation, and rRNA processing. For mobile part (CC) evaluation, they have been enriched in cytosolic ribosome, focal adhesion, extracellular exosome, membrane, and cytosol. for In response to molecular operate (MF) evaluation, they have been enriched in structural constituent of ribosome, rRNA binding, mRNA binding, RNA binding, and cadherin binding. All high 5 BP phrases and high 4 MF phrases have been associated to the ribosome-centered translation course of (Fig. 2D). KEGG pathway enrichment confirmed that these DEPs have been primarily enriched in ribosomal proteins (Fig. 2E).
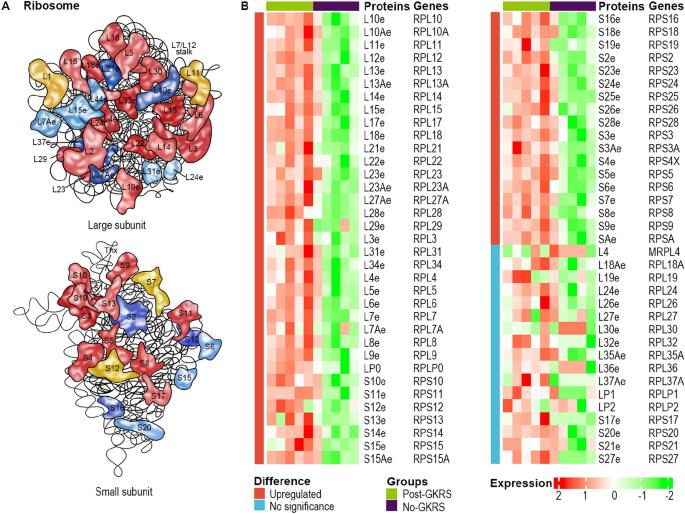
In response to the KEGG pathway hsa03010, 134 proteins have been concerned within the ribosomal pathway (Fig. 3A). On this research, proteomics detected 70 ribosomal proteins out of the 134 ribosomal proteins. As well as, 53 of the 70 (75.7%) ribosomal proteins have been considerably upregulated by GKRS remedy (Fig. 3B). The expression ranges of the remaining 17 (24.3%) ribosomal proteins weren’t considerably altered by GKRS therapy. No ribosomal protein was considerably downregulated. Really, many of the remaining 17 (24.3%) ribosomal proteins have been upregulated by GKRS remedy however with out significance (Fig. 3B). Extra ribosomal proteins may very well be confirmed to extend expression ranges if extra samples are detected. This means that the expression stage of ribosomal proteins is overwhelmingly upregulated after GKRS therapy.
The bulk ribosomal proteins have been upregulated by GKRS remedy. (A) The map of the ribosomal pathway of Homo sapiens (https://www.genome.jp/entry/hsa03010). (B) Heatmaps of the 70 detected ribosomal proteins. The orange column confirmed the 53 ribosomal proteins have been considerably upregulated by γ-knife. The sunshine blue column confirmed no distinction between the two teams. Relative expression ranges have been stained by crimson and inexperienced.
Proteins upregulated by GKRS remedy have been related to a poor prognosis
A earlier research indicated that gamma radiation-upregulated ribosomal proteins may very well be used to enhance radiation resistance by growing the cell’s capability to provide proteins concerned in restoration14. Thus, the expression ranges of ribosomal proteins could also be related to the prognosis of sufferers with lung most cancers. Right here, we explored the connection between the expression ranges of 53 upregulated ribosomal proteins and the general survival of lung most cancers sufferers by analyzing the RNA sequencing dataset of LUAD from The Most cancers Genome Atlas (TCGA).
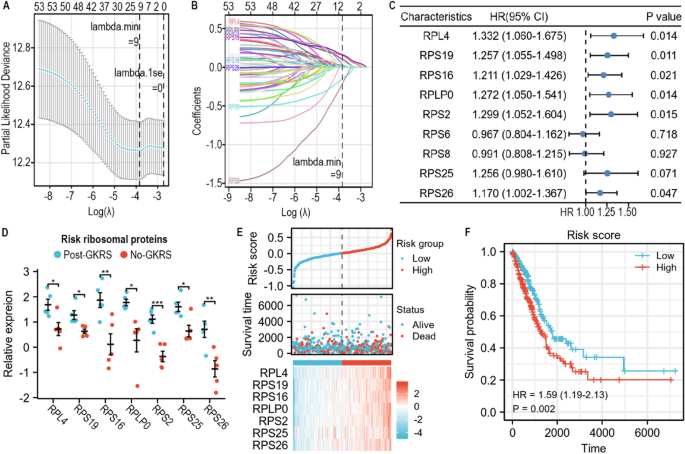
The Lasso coefficient was screened by tenfold cross-validation to acquire potential prognostic proteins. The lambda.min worth was 9 and was used because the cutoff (Fig. 4A). The Lasso variable trajectory diagram confirmed that the potential prognostic proteins have been ribosomal protein (RP) L4 (RPL4), RPS19, RPS16, RPLP0, RPS2, RPS6, RPS8, RPS25, and RPS26 (Fig. 4B). Univariate Cox regression evaluation recommended that the general survival of LUAD sufferers was carefully associated to the expression ranges of RPL4, RPS19, RPS16, RPLP0, RPS2, and RPS26 (Fig. 4C and Desk 2). RPS25, with a p worth of 0.071, was additionally included within the multivariate Cox regression evaluation. Nonetheless, multivariate Cox regression evaluation recommended that none of those proteins have been unbiased danger elements for the prognosis of LUAD sufferers (Desk 2).
Identification of key ribosomal proteins. (A) The lambda.min worth of lasso coefficient was used as cut-off. (B) The potential prognostic proteins have been decided by lasso variable trajectory diagram. (C) Key ribosomal proteins have been recognized by univariate cox regression evaluation with p worth < 0.1. RPL4, RPS19, RPS16, RPLP0, RPS2, RPS26 and RPS25 had shut relation to general survival of LUAD sufferers. (D) The relative expression of the 7 key ribosomal proteins. (E) The connection between danger rating and survival time, expression ranges of seven key ribosomal proteins. (F) The survival curve of danger rating.
Multivariate Cox regression evaluation resulted within the following prognostic system: danger rating = + 0.149378894*Exp(RPL4) + 0.077032501*Exp(RPS19) + 0.009953769*Exp(RPS16) + 0.032681042*Exp(RPLP0) + 0.091708032*Exp(RPS2) − 0.083562796 *Exp(RPS25) + 0.062949989*Exp(RPS26) − 3.043762655. Every case obtained a danger rating. The relative expression ranges of key proteins in BMs are proven in Fig. 4D. All of those proteins have been considerably upregulated by gamma radiation. The prognostic worth of the danger rating was in contrast in response to age, intercourse, major end result of ordinary remedy, distant metastasis, lymph node metastasis, tumor dimension, stage, pack-years smoked, and residual tumor standing. Multivariate Cox regression evaluation recommended that the first end result of ordinary remedy and the danger rating have been unbiased danger elements for the general survival time of LUAD sufferers (Desk 3). With a rise within the danger rating, the mortality price of sufferers elevated and the survival price decreased (Fig. 4E). Log rank survival evaluation additionally confirmed that the danger rating distinguished the general survival time of LUAD sufferers (Fig. 4F).
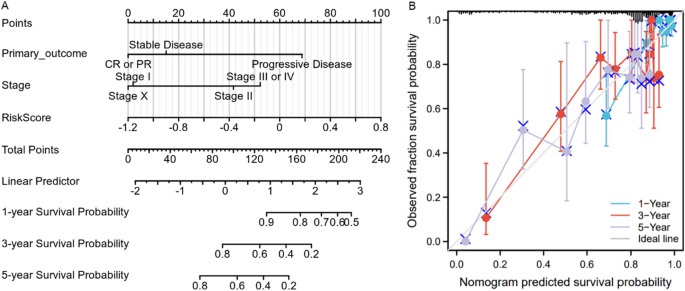
Multivariate Cox regression evaluation additionally recommended that stage was practically an unbiased danger issue for the general survival time of LUAD sufferers (Desk 3). Right here, the danger rating, major end result of ordinary remedy and stage have been used to assemble a nomogram to foretell the 1-year, 3-year, and 5-year survival end result (Fig. 5A). Calibration evaluation was carried out to evaluate the predictive capability of the nomogram. The nomogram calibration plot (Fig. 5B) indicated that the nomogram was nicely calibrated, with imply predicted possibilities for every subgroup near noticed possibilities.