CELF2 deletion is related to myeloid leukemia transformation and enhances HSCs self-renewal
To research the potential position of CELF2 in leukemia, we first analyzed the Pediatric Acute Myeloid Leukemia dataset from the Most cancers Genome Atlas (TCGA) (http://www.cbioportal.org/). We discovered that CELF2 deletion occurred in 9.4% of acute myeloid leukemia (AML) sufferers (Fig. S1A) and the survival time of AML sufferers with CELF2 deletion was considerably shorter than that of the others (Fig. S1B). Curiously, MLL associated genetic alteration accompanied with CELF2 deletion accounted for six.38% of all of the 94 AML sufferers (Desk S1). Extra mRNA expression analyses of CELF2 confirmed the expression is highest in regular myeloid cells in peripheral blood (PB) cells utilizing information from human leukemia databases (GSE42519 and GSE13159, http://servers.binf.ku.dk/bloodspot), whereas it decreased in numerous subtypes of AMLs (Fig. S1C). We additional confirmed that CELF2 mRNA degree was considerably decrease in major AML cells from sufferers’ complete bone marrow (BM) when put next with the mononuclear cells from wholesome donor BM (Fig. S1D). These information recommended that CELF2 could play an necessary position in leukemogenesis in AML sufferers.
To additional dissect the position of CELF2 in hematopoiesis and leukemogenesis, we generated Celf2fl/fl mice (Celf2 WT) (Fig. S1E) and crossed them with Vav1-Cre mouse line to generate hematopoietic-specific Celf2-poor mice (Vav1-Cre; Celf2fl/fl, will likely be referred to Celf2 KO) (Fig. S1F, G). We discovered that the burden of spleen was considerably elevated in Celf2 KO mice when put next with the controls, whereas the physique weight of Celf2 KO and management mice was comparable (Fig. 1A). The histopathologic examination confirmed no dysplasia in bone marrow (BM) and spleen in Celf2 KO mice (Fig. 1B). Celf2 KO mice had increased white blood cell (WBC) counts and monocytes in PB than that of Celf2 WT mice (Fig. 1C). Circulation cytometric evaluation of BM from two teams of mice confirmed that the share of HSCs was comparable, whereas the share of HPCs and GMPs/CMPs was considerably elevated in Celf2 KO mice as compared with the controls respectively (Fig. 1D, E). Absolutely the variety of HPCs, HSCs, CMPs, CMPs and MEPs from CELF2 KO mice was equal to that of controls (Fig. S1H). Moreover, we used SLAM markers to characterize the LT-HSCs and located absolutely the variety of LT-HSCs from CELF2 KO mice was considerably elevated when put next with that of controls (Fig. S1I). Though the share of myeloid cells was increased in BM of Celf2 KO mice than that of controls (Fig. 1F), the differentiation of myeloid cells was regular in PB and BM of Celf2 KO mice by Wright-Giemsa staining assay (Fig. 1G). These outcomes indicated that lack of Celf2 will increase the myeloid progenitor cells pool and results in the growth of myeloid cells mildly in vivo.
A The load of mice and the burden of spleen from 8 weeks Celf2fl/fl or Celf2 KO mice (n = 5). B H&E staining of BM and spleen in Celf2fl/fl or Celf2 KO mice at 8 weeks. C Blood rely evaluation within the PB of Celf2fl/fl or Celf2 KO mice at 8 weeks for white blood cells (WBC), monocytes (MON) (n = 5). D Consultant circulation cytometry plot and frequencies of HPCs and HSCs within the bone marrow of Celf2fl/fl or Celf2 KO mice at 8 weeks (n = 5). E Consultant circulation cytometry plot and frequencies of the GMPs, CMPs and MEPs in complete BM cells of Celf2fl/fl or Celf2 KO mice at 8 weeks (n = 5). F Wright-Giemsa stain of peripheral blood and bone marrow in Celf2fl/fl or Celf2 KO mice at 8 weeks. G Percentages of various mature lineage cells in BM of Celf2fl/fl or Celf2 KO mice at 8 weeks (n = 5). H Whole BM cells from Celf2fl/fl or Celf2 KO mice had been transplanted into lethally irradiated recipient mice (CD45.1) along with equal numbers of CD45.1 competitor BM cells. Circulation cytometry evaluation of the contribution of Celf2fl/fl (CD45.2+) and Celf2 KO (CD45.2+) donor-derived cells within the PB of recipient mice after aggressive BMT in major transplantation assay (left panel) and secondary transplantation assay (proper panel) (n = 7). I The chances of Celf2fl/fl and Celf2 KO donor-derived myeloid cells within the PB of recipient mice after aggressive BMT (n = 7). J Share of donor-derived totally different mature lineage cells compartments within the BM of recipients at 16 weeks after aggressive BMT (n = 7). Okay Share of donor-derived GMPs, CMPs and MEPs compartments in spleen of recipients at 16 weeks after aggressive BMT (n = 7). L Share of donor-derived LSK cells at 16 weeks after aggressive BMT (n = 7). The information are offered because the imply ± SD and one-way ANOVA, **P < 0.01; ****P < 0.0001.
To additional probe the potential position of Celf2 in regulating the operate of HSCs, we carried out aggressive transplantation assay by transplanting BM cells from Celf2 KO or Celf2 WT mice along with CD45.1+ BM cells into lethally irradiated CD45.1+ recipient mice. We examined the frequency of reconstituted cells (CD45.2+) in PB each 4 weeks after transplantation and located that the chimerism of Celf2 KO cells (CD45.2+) in PB was considerably increased than the controls (Fig. 1H, left panel). Moreover, in our serial aggressive transplantation assay, whereas the chimerism of Celf2 KO cells (CD45.2+) in PB was twice that of the management in major (1°), the change was decreased in secondary (2°) transplantation (Fig. 1H, proper panel), suggesting that the aggressive benefit of Celf2 KO HSCs could also be diminished throughout serial transplantations. Notably, the myeloid cells derived from Celf2 KO BM cells had been elevated (Fig. 1I). We additionally discovered that the share of T cells derived from Celf2 KO BM cells had been decrease than that of controls (Fig. S1J), whereas the share of B cells derived from Celf2 KO BM cells had been increased than that of controls (Fig. S1K). The recipient mice had been sacrificed at 4 months after transplantation, and the chances of donor-derived Celf2 KO CMPs/GMPs in spleen and myeloid cells in BM had been discovered to be elevated in recipient mice (Fig. 1J, Okay). The proportion of donor-derived Celf2 KO CMPs/GMPs in BM was equal to that of controls (Fig. S1L). Notably, the variety of donor-derived Celf2 KO LSK+ cells was a lot increased within the BM of recipient mice after four-month transplantation, when put next with that of the controls (Fig. 1L). In abstract, these outcomes demonstrated that Celf2 deletion enhances hematopoietic reconstitution however doesn’t result in malignant hematopoiesis.
Celf2 deficiency lowers the edge of malignant transformation of hematopoietic progenitor cells to leukemia
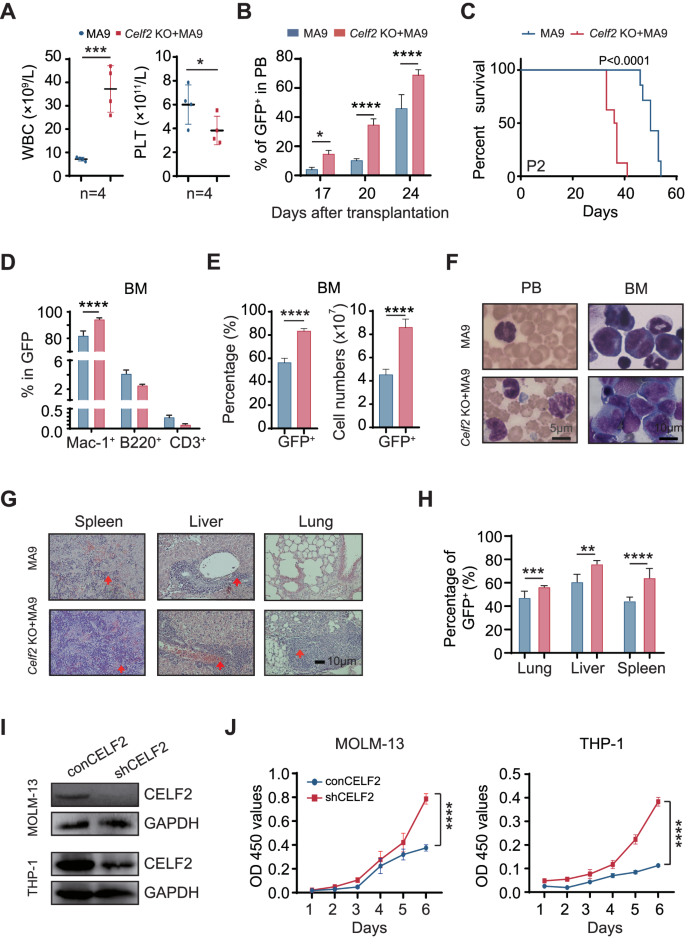
In distinction to many different leukemia-associated mutations, MLL-fusions are highly effective oncogenes that might rework each hematopoietic stem cells and dedicated progenitor cells. To judge the potential synergistic motion of Celf2 loss and MLL-fusion within the leukemia-initiating occasion in AML growth, we transduced hematopoietic progenitor cells from Celf2 KO and Celf2 WT mice with retrovirus expressing MA9 (Fig. S2A and Supplemental methodology). On this mannequin, Celf2 KO + MA9 mice confirmed extra aggressive leukemia phenotypes than Celf2 WT + MA9 mice, together with increased WBCs and decrease PLT in PB (Fig. 2A). Furthermore, the share of leukemia cells from 17 to 24 days after transplantation confirmed accelerated leukemia growth in Celf2 KO + MA9 mice (Fig. 2B), resulting in markedly accelerated mortality on this group (Celf2 KO + MA9 mice median survival = 36 days; MA9 mice median survival = 50 days; log-rank check P < 0.0001; Fig. 2C). Additional evaluation confirmed that the GFP+ leukemia cells in these two teams had been primarily myeloid cells (Mac-1+) (Fig. 2D). The proportion and absolute variety of GFP+ leukemia cells had been considerably elevated in BM of Celf2 KO + MA9 mice, when put next with that of MA9 mice (Fig. 2E). Wright-Giemsa staining confirmed that the counts of irregular leukemia cells was elevated in PB and BM of Celf2 KO + MA9 mice when put next with that of MA9 mice (Fig. 2F). The proportion of GFP+ leukemia cells and the diploma of extramedullary infiltration had been additionally elevated in Celf2 KO + MA9 mice than that in MA9 mice, together with within the spleen, liver and lung (Fig. 2G, H).
A Full blood rely evaluation of peripheral blood (PB), white blood cells (WBC) and platelets (PLT) in MA9 or Celf2 KO + AML mice (n = 5). B The proportion of GFP+ cells in PB after transplantation (n = 7). C Kaplan–Meier survival evaluation of MA9 or Celf2 KO + MA9 mice in secondary transplant (n = 7). The information are offered because the imply ± SD and one-way ANOVA, **P < 0.01; ****P < 0.0001. D The lineage commitments of the transduced HSPCs had been evaluated by analyzing the chances of myeloid cells, B lymphocytes and T lymphocytes throughout the GFP+ cells. The information for the final time level had been collected when the mice had been euthanized (n = 5). E The proportion and absolute variety of GFP+ cells in BM of MA9 or Celf2 KO + AML mice (n = 5). F Wright-Giemsa stain of PB and BM. G H&E staining of spleen, liver and lung. H The proportion of GFP+ cells in spleen, liver and lung from recipient mice at finish stage (n = 5). I Western blotting exhibiting CELF2 KD effectivity in MOLM-13 and THP-1 leukemia cells after transduction with shRNA lentiviruses focusing on CELF2. J Cell proliferation within the indicated cell strains after transduction with lentivirus expressing management or CELF2-specific shRNA. Panel I, J reveals 1 consultant experiment of a minimum of 3 impartial experiments.
To research whether or not the CELF2 deletion is functionally related to human myeloid leukemia growth, we knocked down (KD) the CELF2 expression in MOLM-13 and THP-1 (each comprise MLL-AF9 mutation) cell strains (Fig. 2I), and located that CELF2 KD elevated the expansion of MOLM-13 and THP-1 cells (Fig. 2J), decreased the apoptosis of MOLM-13 and THP-1 cells (Fig. S2B). After we overexpressed (OE) the CELF2 in MOLM-13 cells (Fig. S2C), we discovered that CELF2 OE considerably decreased the expression of P-AKT and inhibited the expansion of MOLM-13 cells in vitro (Fig. S2C, D). Taken collectively, these outcomes demonstrated that CELF2 loss might considerably speed up myeloid leukemia development in vitro and decrease the edge of MLL-AF9-induced AML growth in vivo.
Celf2 deficiency potentiates the potential of leukemia stem cells in MA9-induced AML
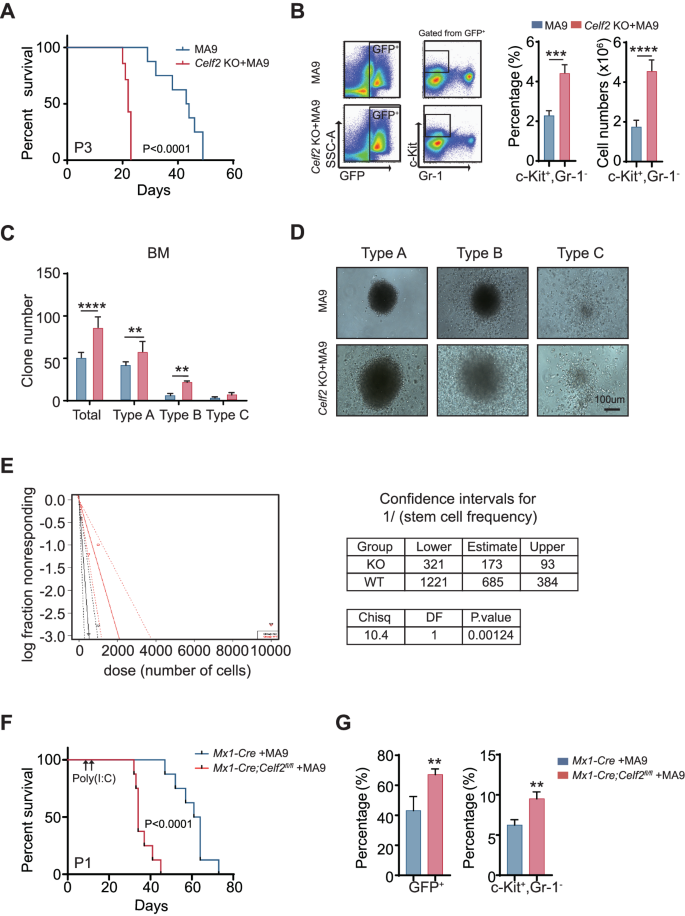
To find out how Celf2 deletion accelerates leukemia initiation, we serially transplanted the equal numbers of Celf2 KO + MA9 or MA9 leukemia cells into the recipient mice. The mice transplanted with Celf2 KO + MA9 cells had extra leukemic cells within the PB, BM and extramedullary organs than the controls three weeks after transplantation (Fig. S3A). Notably, over 40% of MA9 mice survived past 40 days after transplantation, whereas all Celf2 KO + MA9 mice died inside 22 days after transplantation (Fig. 3A), suggesting that Celf2 deletion could enhance the exercise of LSCs.
A Kaplan–Meier survival evaluation of MA9 or Celf2 KO + AML mice in tertiary transplant (n = 8). The information are offered because the imply ± SD and one-way ANOVA, **P < 0.01; ****P < 0.0001. B The proportion of c-Package+Gr-1− cells from BM in MA9 or Celf2 KO + AML mice (n = 5). Colony formation assay (C), and consultant colony photos (D) of murine leukemia cells. Cells had been cultured in MethoCult M3434 for 7 days. E The frequency of LSCs was decided by serial dilution and aggressive transplantation assay. F Kaplan–Meier survival evaluation of Mx1-Cre + MA9 or Mx1-Cre;Celf2fl/fl + AML mice post-poly(I:C) injection (n = 8). G The proportion of GFP+ (left panel) or c-Package+Gr-1− (proper panel) cells from BM in Mx1-Cre + MA9 or Mx1-Cre;Celf2fl/fl + MA9 AML mice post-poly(I:C) injection (n = 4).
The c-Package+Gr-1− leukemia cells had been usually outlined as LSCs in AML mouse mannequin [18]. We discovered that the quantity and the share of c-Package+Gr-1− leukemia cells had been elevated in BM of Celf2 KO + MA9 mice when put next with that of MA9 mice (Fig. 3B). Then, we carried out the colony-forming assay and evaluated the self-renewal capability of Celf2 KO + MA9 LSCs in vitro. Three kinds of colonies had been outlined primarily based on the literature: kind A colonies had a compact heart, kind B colonies had a dense heart surrounded by a halo of migrating cells, and sort C colonies consisted of diffuse differentiating cells [19]. The outcomes revealed that Celf2 KO + MA9 leukemia cells gave rise to extra colonies with bigger measurement than MA9 leukemia cells, particularly kind A colonies (Fig. 3C, D), indicating that Celf2 deletion elevated the variety of LSCs and colony-forming exercise, in addition to leukemic cell development potential. We additionally discovered that the variety of Celf2 KO + MA9 cells was considerably elevated than the variety of MA9 cells (Fig. S3B). According to extra LSCs within the Celf2 KO + MA9 cell inhabitants, excessive limiting dilution transplantation assays demonstrated a marked enhance in leukemia-initiating cell frequency in Celf2 KO + MA9 AML cells when put next with MA9 AML cells (Fig. 3E).
Subsequent, we evaluated the consequence of Celf2 loss within the upkeep of established AML leukemia. Mx1-Cre or Mx1-Cre; Celf2fl/fl mouse cells had been remodeled by retrovirus containing MA9 and transplanted to lethally irradiated recipients. After 10 days, Cre expression and Celf2 gene excision had been induced by three intraperitoneal injections of poly(I:C) given each different day. The deletion of Celf2 post-poly(I:C) induction was confirmed by PCR evaluation and Western Blotting after BM aspiration (Fig. S3C-D). Deletion of Celf2 considerably enhanced leukemia development in MA9 mice (Fig. 3F). According to the phenotypes of Vav1-Cre; Celf2fl/fl + MA9 mice, the mice transplanted with Mx1-Cre; Celf2fl/fl + MA9 cells had extra leukemic cells in BM than the controls two weeks after transplantation (Fig. 3G, left panel). Amongst them, LSCs (c-Package+Gr-1−) had been additionally considerably elevated (Fig. 3G, proper panel). Taken as a complete, our outcomes confirmed that Celf2 loss enhanced the frequency and self-renewal capability of MA9-transformed LSCs, accelerated the incidence and the event of MA9-driven AML.
Celf2 deficiency prompts leukemia associated pathways
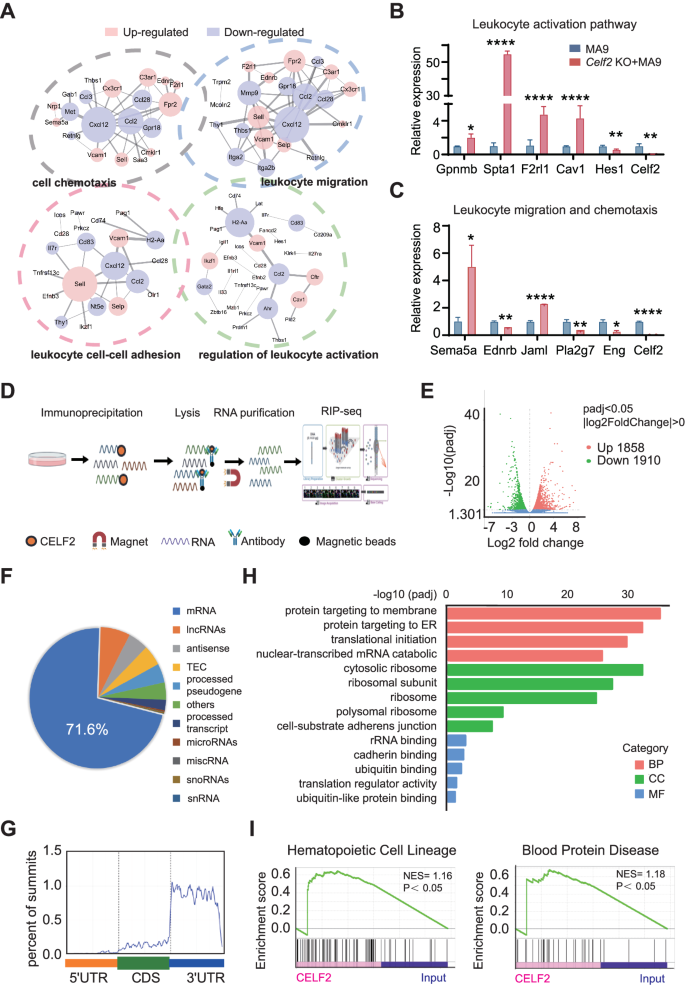
To dissect the underlying molecular mechanism that drives the improved oncogenic potential in Celf2 KO + MA9 cells, we carried out international gene expression evaluation of leukemia cells from Celf2 KO + MA9 mice and the management MA9 mice by RNA-Seq. 1569 genes had been discovered to be differentially expressed in leukemia cells with the presence or absence of Celf2 (p < 0.05, |fold change| > 2) (Fig. S4). The differentially expressed genes might be categorized primarily into 4 practical teams, together with genes encoding merchandise with cell chemotaxis, leukocyte migration, leukocyte cell-cell adhesion and leukocyte activation (Fig. 4A), suggesting that a wide range of molecular and mobile processes have been altered in Celf2 KO + MA9 cells. The consultant prime fold-changed genes had been validated by Realtime-PCR, together with these concerned in leukocyte activation (Gpnmb, Spta1, F2rl1, Cav1 and Hes1) and leukocyte migration (Sema5a, Ednrb, Jaml, Pla2g7 and Eng) (Fig. 4B, C). Our information recommend that lack of Celf2 seemingly enhances the self-renewal and survival of Celf2 KO + MA9 cells by means of a number of molecular pathways that contribute to the MA9-induced leukemia.
A Gene ontology (GO) enrichment of 4 distinct signaling communities amongst RNA-Seq of MA9 or Celf2 KO + AML mice. Ranges of the pathway-related genes mRNA (B: leukocyte activation pathway; C: leukocyte migration and chemotaxis pathway) in MA9 or Celf2 KO + AML leukemia cells had been analyzed utilizing RT-PCR. D Define of the experimental strategies for the CELF2 RIP-seq in leukemia cells. E Volcano plot of differentially enriched genes in RIP-Seq. The values of X and Y axes within the volcano plot are because the fold change (log2 remodeled) and P adj (−log10 remodeled) between two teams, respectively. Purple/Inexperienced dots point out 2-fold change differentially enriched genes with statistical significance. Blue dots point out non-differentially enriched genes. F Percentages of varied RNA species for RIP-seq of leukemia cells. G GO enrichment evaluation of phrases enriched in upregulated genes in RIP-Seq. H Metagene profiles of enrichment of CELF2-binding websites throughout mRNA transcriptome. CDS, coding sequence. I Enrichment plots from gene set enrichment evaluation (GSEA) primarily based on RIP-Seq information (left panel: hematopoietic cell lineage associated genes; proper panel: blood protein illness associated genes) (normalized enrichment rating [NES] is inferred from permutations of the gene set and the false discovery fee [FDR]).
To establish the molecular effectors that straight mediated by CELF2 in myeloid leukemia, we profiled the CELF2-binding RNAs at transcriptome degree. We overexpressed 3xFlag-HA-CELF2 and carried out RIP and high-throughput sequencing (RIP-seq) in K562 cells (Fig. 4D). Among the many 1858 CELF2-dependent transcripts recognized, 71.6% had been mRNAs (Fig. 4E, F), and primarily had been within the 3’ untranslated area (UTR) stem loop areas (Fig. 4G). GO evaluation confirmed that CELF2-bound RNAs had been enriched in varied organic capabilities, resembling membrane protein localization associated to migration and adhesion, RNA degradation associated to three’UTR, translational initiation (Fig. 4H). GSEA evaluation of those targets confirmed vital enrichment in pathways associated to hematopoietic cell lineage and blood protein ailments (Fig. 4I), in step with the important position of CELF2 in hematopoiesis and leukemogenesis. Our RIP-seq outcomes indicated that CELF2 could straight bind to a number of goal mRNAs that take part in hematopoiesis and leukemogenesis
Deletion of CELF2 enhances FAT10 mRNA stability which prompts the mTOR pathway, and promotes AML development
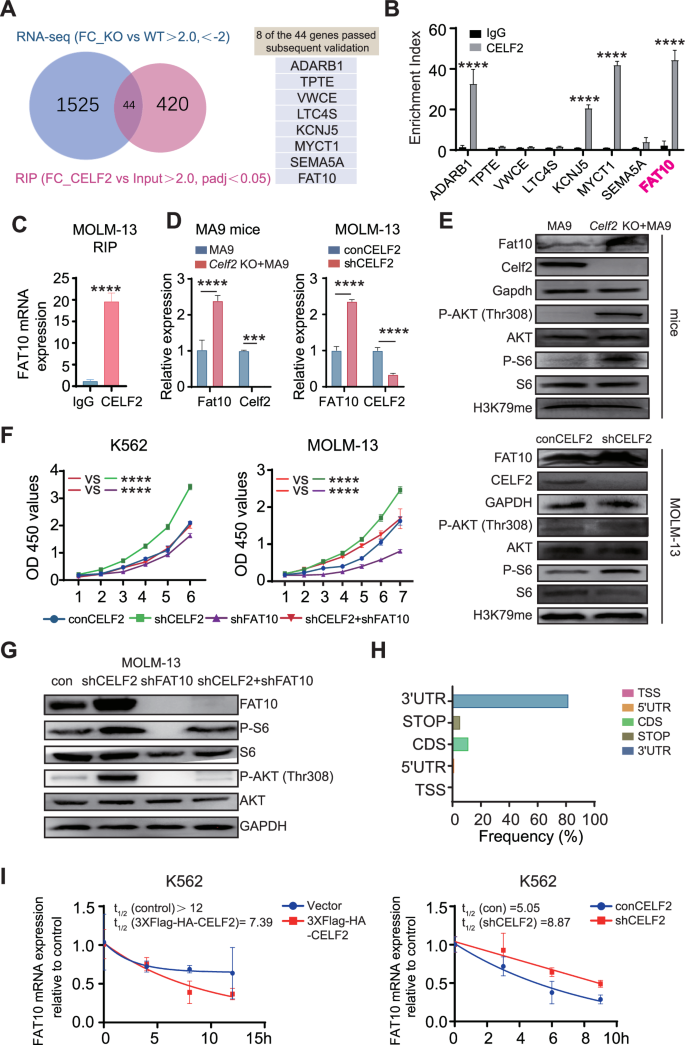
To particularly outline the downstream alerts of CELF2 which can be crucial for leukemia development, we analyzed built-in information from RIP-Seq and RNA-Seq to establish the goal mRNA of CELF2. Utilizing the standards of padj < 0.05 and fold-change worth > 2, we recognized 464 mRNA targets of the CELF2 protein in RIP-Seq and 647 genes that had been differentially expressed above the edge degree in RNA-seq of leukemia cells from Celf2 KO + MA9 mice and the management MA9 mice. 44 putative mRNA targets had been obtained by way of the built-in information (Fig. 5A). Of those, utilizing qRT-PCR, 8 genes had been discovered to be each enriched in RIP and dysregulated by CELF2 in AML cells (Fig. 5B). FAT10 (Human Leukocyte Antigen-F adjoining transcript 10) mRNA was discovered to be extremely enriched by CELF2 RIP, and its mRNA and protein had been considerably upregulated in Celf2 KO AML mice and cells (Fig. 5B–E). We noticed that FAT10 was extremely expressed in AML affected person samples when put next with regular people (Fig. S5A). Since FAT10 has been reported to straight promote most cancers cell proliferation in varied cancers by activating the AKT-mTORC1 signaling pathway [20], we examined the degrees of FAT10, P-AKT (Thr308), P-S6 and H3K79me in Celf2 KO + MA9 AML mouse cells and CELF2 KD human MOLM-13 and K562 myeloid leukemia cells. We discovered that the degrees of FAT10, P-AKT (Thr308) and P-S6 had been considerably elevated in each murine and human cells when put next with Celf2 WT + MA9 AML cells and CELF2 WT human MOLM-13 and K562 myeloid leukemia cells respectively, whereas the extent of H3K79me was equal in Celf2 KO + MA9 and MA9 AML cells (Figs. 5E and S5B). Curiously, the extent of P-AKT in Celf2 KO HSCs and management HSCs measured by circulation cytometry didn’t present considerably elevated in Celf2 KO HSCs when put next with that of controls (Fig. S5C). We additional knocked down FAT10 in MOLM-13 and K562 cells, noticed that FAT10 KD diminished the P-AKT (Thr308) and P-S6 degree (Fig. S5D). Our outcomes thus demonstrated that FAT10 expression discount might inhibits AKT pathway activation in AML.
A Venn diagram illustrating the overlap of 44 mRNA targets of CELF2 recognized utilizing the RIP-seq and RNA-seq evaluation (left panel); eight transcripts handed follow-up validation (proper panel). B RT-PCR evaluation of eight transcripts obtained from RIP with anti-CELF2 and IgG management antibodies in K562 cells. C RT-PCR evaluation of FAT10 with anti-CELF2 and IgG management antibodies in MOLM-13 cells. D Ranges of the Fat10 (FAT10) mRNA in MA9 mice (left panel) and MOLM-13 (proper panel) had been analyzed utilizing RT-PCR. E Ranges of pathway-related proteins in Celf2 (CELF2) knockdown leukemia cells (higher panels: mice; decrease panels: MOLM-13) had been analyzed by Western blot. F Cell proliferation within the indicated cell strains after transduction with lentivirus expressing management, CELF2-specific shRNA, FAT10-specific shRNA or CELF2-specific shRNA and FAT10-specific shRNA. G Ranges of pathway-related proteins in MOLM-13 after transduction with lentivirus expressing management, CELF2-specific shRNA, FAT10-specific shRNA or CELF2-specific shRNA and FAT10-specific shRNA. H The distribution of CELF2-binding peaks inside totally different gene areas recognized by FLAG-CELF2 RIP-seq in K562 cells. I After remedy with 10 mg/mL Actinomycin-D, complete RNA was extracted at particular time. The half-lives of FAT10 mRNA in CELF2-overexpressing K562 cells and CELF2 knockdown K562 cells had been measured. Panels B–F, H present 1 consultant of a minimum of 3 impartial experiments.
To additional examine whether or not CELF2 regulates AKT-mTORC1 signaling pathway by way of FAT10, FAT10 was knocked down in CELF2 KD MOLM-13 and CELF2 KD K562 cells. We discovered that decreased FAT10 expression inhibited the over-growth of CELF2 KD myeloid leukemia cells, reversed the impact of CELF2 deletion (Fig. 5F). Notably, the phosphorylation ranges of P-AKT (Thr308) and P-S6 had been considerably diminished in FAT10 KD MOLM-13 and FAT10 KD K562 cells (Figs. 5G and S5E). We additional knocked down FAT10 (FAT10 KD) in CELF2 KD MOLM-13 cells and transplanted the cells to BALB/c nude mice to find out the cell proliferation in vivo. We discovered that FAT10 KD might inhibit the expansion of CELF2 KD MOLM-13 cells when put next with the management group (Fig. S5F).
We additional analyzed the connection between CELF2 and FAT10 in AML sufferers and regular people from dataset (LAML tumor and LAML regular) in GEPIA (Gene Expression Profiling Interactive Evaluation) (http://gepia.cancer-pku.cn/element.php?gene=CELF2). We discovered that the expression of CELF2 is negatively correlated with the expression FAT10. The expression of FAT10 is excessive in BM cells with CELF2 low expression (Fig. S5G). Taken collectively, these outcomes demonstrated that lack of CELF2 accelerated the myeloid leukemia cell proliferation by activating the FAT10-AKT-mTORC1 signaling pathway.
To research how CELF2 regulates FAT10, we examined the situation of the mRNA compete-derived consensus motif and located that CELF2 was extremely enriched in 3′UTRs of focusing on mRNA (Fig. 5H). We additional analyzed the uncooked information of RIP-seq and the IGV peaks for the binding websites of CELF2 on FAT10 (Fig. S5H). Our outcomes confirmed that CELF2 is normally enriched within the FAT10 3′UTR area. Moreover, we discovered that over-expression of CELF2 considerably promoted the degradation of FAT10 mRNA in K562 cells following actinomycin-D (10 μg/mL) remedy, whereas the FAT10 transcripts exhibited longer half-lives in CELF2 KD K562 cells (Fig. 5I). These outcomes recommended that CELF2 straight fine-tunes the transcription of FAT10, thus inhibits the FAT10-AKT-mTORC1 signaling pathway in myeloid leukemia cells.
As well as, The AU-rich components (AREs) in 3’UTRs are probably the most prevalent cis-destabilizing RNA motifs [21, 22]. ARE-containing mRNA transcripts are thought-about to be inherently unstable, and are subjected to dynamic decay for mRNA circulation [21]. Earlier research indicated that CELF2 might regulate goal gene expression by binding to AREs within the 3′UTR of its goal mRNAs [23,24,25]. By analyzing the sequence of FAT10 3’UTR area, we discovered three AREs (Fig. S6A). We constructed the luciferase (Fluc) reporter that was inserted with totally different AREs of FAT10 and examined the Fluc exercise in CELF2 KD K562 cells and controls (Fig. S6B). We discovered that CELF2 KD induced a considerably enhance in Fluc exercise of FAT10 reporter (Fig. S6C), indicating that CELF2 are seemingly binding to the final AREs in 3′UTR of FAT10. In abstract, we recognized the three′UTR of FAT10 mRNA incorporates ARE areas as a possible practical CELF2-binding web site, which can contribute to the decay of FAT10 mRNA.
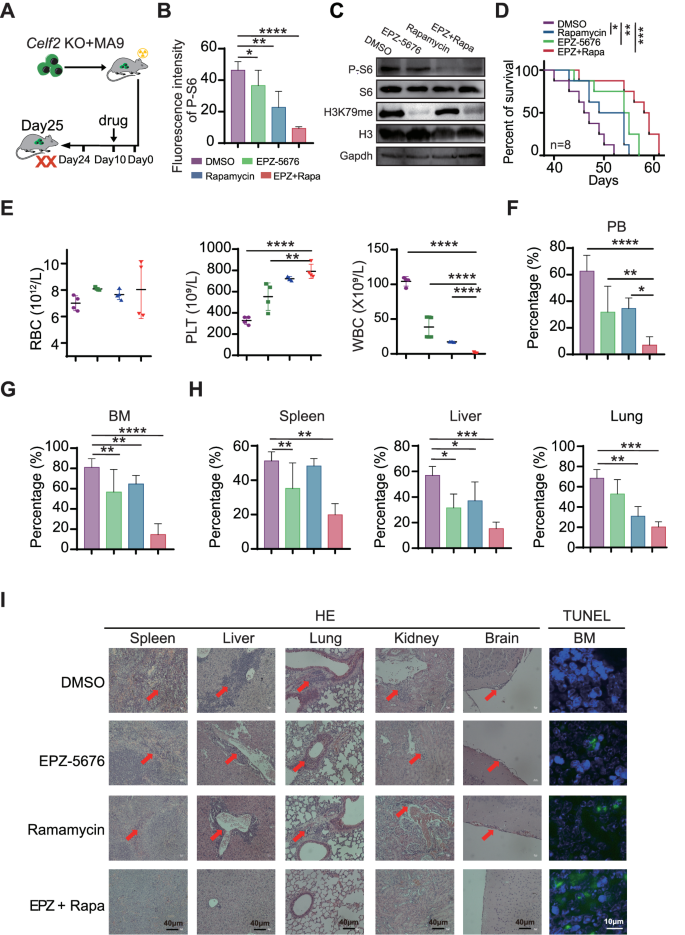
Mixed remedy with EPZ-5676 and Rapamycin reveals synergistic anti-tumor results in Celf2 KO + MA9 mouse mannequin
Since FAT10-mTORC1 signaling pathway performs a central position within the pathogenesis of Celf2 KO + MA9 AML, we sought to find out whether or not twin inhibition of mTORC1 and MA9 pathways could be useful for Celf2 KO + MA9 AML mice. Rapamycin is a mTOR inhibitor whereas pinometostat (EPZ-5676) is a potent DOT1L histone methyltransferase inhibitor which might successfully inhibit MLL-fusion goal gene expression in vivo. Celf2 KO + MA9 mice had been handled with both placebo, single EPZ-5676, single rapamycin, or mixed EPZ-5676 and rapamycin (Fig. 6A). We discovered that the degrees of P-S6 and H3K79me in Celf2 KO + MA9 cells had been considerably decreased, whereas the protein ranges of S6 and H3 had been equal in BM cells (Figs. 6B, C and S7A). The survival time of the mice handled with EPZ-5676+rapamycin was considerably longer than that of mice handled with different remedy (Fig. 6D). The counts of PLT had been elevated, whereas the rely of WBCs was decreased within the PB of mice handled with EPZ-5676+rapamycin when put next with that of the mice handled with single EPZ-5676 or rapamycin solely (Fig. 6E). Moreover, the chances of GFP+ leukemia cells within the PB, BM, spleen, liver and lung had been decreased in mice handled with EPZ-5676+rapamycin when put next with these in different teams (Fig. 6F, H). HE staining confirmed that the diploma of leukemia cell infiltration within the spleen, liver, lung, kidney and mind was diminished in mice handled with EPZ-5676+rapamycin when put next with different teams (Fig. 6I, HE panels). Additionally, immunohistochemical staining of Ki67, a marker of cell proliferation, confirmed considerably diminished staining within the EPZ-5676+rapamycin-treated group when put next with the opposite teams (Fig. S7B), whereas TUNEL (TdT-mediated dUTP nick-end labeling) staining, an indicator of apoptotic cells, was proven to be elevated in mice handled with EPZ-5676+rapamycin when put next with the opposite teams (Fig. 6I, TUNEL panels). Thus, our outcomes confirmed that mixed remedy with EPZ-5676 and rapamycin might scale back Celf2 KO + MA9 leukemia burden and extend the survival time of Celf2 KO + MA9 AML mice by inhibiting each mTORC1 pathway and MLL-fusion goal gene expression.
A Schematic illustration of various drug therapies in Celf2 KO + MA9 mice. B Phospho-flow cytometry was used to measure the phosphorylation degree of S6 in leukemia cells from Celf2 KO + MA9 mice handled with placebo, single EPZ-5676, single rapamycin, or mixed EPZ-5676 and rapamycin by means of tail vein injection (n = 5). C Western blotting was used to evaluate the expression of P-S6, S6, H3K79me, H3K79 in leukemia cells from Celf2 KO + MA9 mice handled with totally different medication. D Kaplan–Meier survival curves of Celf2 KO + MA9 mice handled with totally different medication (n = 8). E The counts of RBC, PLT and WBC in PB by routine blood assessments. The proportion of GFP+ leukemia cells in PB (F) and BM (G) from Celf2 KO + MA9 mice handled with totally different medication (n = 5). H The proportion of GFP+ leukemia cells in organs (spleen, liver, lung) from Celf2 KO + MA9 mice handled with totally different medication (n = 5). I Histological staining for hematoxylin and eosin (HE) in spleen, liver, lung, kidney and mind (magnification, ×40), and TUNEL staining in BM (inexperienced represents TUNEL, blue represents DAPI, magnification, ×60), from Celf2 KO + MA9 mice handled with totally different medication.