Pan-cancer convolutional neural networks for tumor/regular classification
We developed a CNN structure to categorise slides from TCGA by tumor/regular standing, utilizing a neural community that feeds the final totally linked layer of an Inception v3-based CNN pretrained on ImageNet into a completely linked layer with 1024 neurons. This structure is depicted in Fig. 1a, and a associated structure for mutation classification (described in sections beneath) is proven for comparability in Fig. 1b. The 2 last totally linked layers of the tumor/regular CNN had been educated on tiles of measurement 512 × 512 from WSIs. Because of inadequate FFPE regular WSIs in TCGA, for this job, we solely used flash-frozen samples. We educated this mannequin individually on slides from 19 TCGA most cancers sorts having numbers of slides starting from 205 to 1949 (Fig. 2a). In all, 70% of the slides had been randomly assigned to the coaching set and the remaining had been assigned to the take a look at set. To deal with the information imbalance downside28, the bulk class was undersampled to match the minority class.
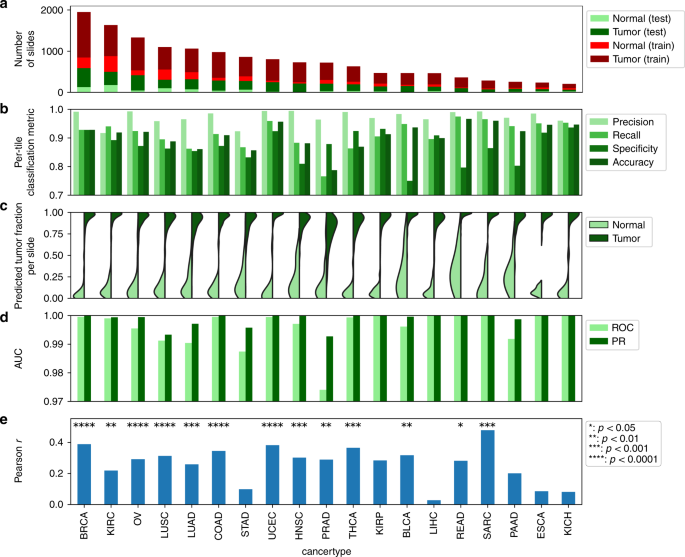
a Numbers of tumor and regular slides in take a look at and coaching units. b Per-tile classification metrics. Tile-level take a look at set sizes are offered in Supplementary Information 3. c Fraction of tiles predicted to be tumor inside every slide. d Per-slide AUC values for tumor/regular classification for ROC and precision-recall curve (PR). e Pearson correlation coefficients between predicted and pathologist analysis of tumor purity. P values are primarily based on the permutation take a look at of the dependent variable after Bonferroni correction throughout all most cancers sorts. Uncooked and adjusted P values are offered in Supplementary Information 4.
Determine 2b reveals the classification outcomes. We used a naive coaching method such that, after removing of background areas, all tiles in a traditional picture are assumed regular and all tiles in a tumor picture are assumed tumor. The CNN precisely classifies take a look at tiles for many tumor sorts (accuracy: 0.91 ± 0.05, precision: 0.97 ± 0.02, recall: 0.90 ± 0.06, specificity: 0.86 ± 0.07. Imply and normal deviation calculated throughout most cancers sorts). We subsequent examined the fraction of tiles categorized as tumor or regular inside every slide. The fractions of tiles matching the slide annotation are 0.88 ± 0.14 and 0.90 ± 0.13 for regular and tumor samples, respectively (Fig. 2c) (imply and normal deviation calculated from all most cancers sorts pooled collectively). These fractions are excessive in nearly all slides, and the tumor-predicted fraction (TPF) is considerably completely different between tumors and normals (P < 0.0001 per-cohort comparability of tumor vs. regular, Welch’s t take a look at). We additionally carried out the classification on a per-slide foundation. To do that, we used the TPF in every slide as a metric to categorise it as tumor or regular. This method yielded extraordinarily correct classification outcomes for all most cancers sorts (Fig. 2nd, imply AUC ROC = 0.995, imply PR AUC = 0.998). Confidence intervals (CI) of per-slide predictions are given in Supplementary Fig. 1a (additionally see “Strategies”). The CI decrease certain on all classification fashions was above 90%, with most cancers sorts having fewer slides or imbalanced take a look at information tending to have bigger CIs. These outcomes point out that our community can efficiently classify WSIs as tumor/regular throughout many various most cancers sorts. These outcomes had been for slide-level take a look at/prepare splits of the information, however splitting on the affected person degree as a substitute had little impact on classification accuracy (see “Strategies” and Supplementary Fig. 1b). Most misclassification was for adjoining regular slides with unexpectedly giant predictions for TPF. Handbook pathology evaluate indicated that such slides typically undergo from poor high quality, tissue folding, or extreme tissue injury associated to freezing (e.g., see Supplementary Fig. 3).
We subsequent investigated if TPF correlates with tumor purity, that’s, slides with increased tumor purity are inclined to have bigger TPFs and vice versa. We discovered important optimistic correlations between TPF and TCGA pathologist-reported purity (“common share of tumor cells”) within the majority of most cancers sorts (Fig. 2e), with bigger most cancers units are inclined to have extra important p values (e.g., BRCA: P = 5e-17). The distributions of TPF had been systematically increased than the pathologist annotations (Supplementary Fig. 2), although this distinction might be partially reconciled by the truth that TPF is predicated on the neoplastic space whereas the pathologist annotation is predicated on cell counts. Tumor cells are bigger than stromal cells and cut back nuclear density. Whereas TPF and purity are clearly associated, the reasonable magnitudes of correlations point out that intraslide enhancements might be made. A notable limitation is the coaching assumption that tiles in a slide are both all tumor or all regular, as intraslide pathologist annotations will not be offered by TCGA. Moreover, pathologist assessments of tumor purity have non-negligible variability29 that will have an effect on correlations. For comparability, we additionally calculated the correlation of TCGA pathologist-reported purity with the genomics-inferred purity measures ABSOLUTE30 and InfiniumPurify31 in BRCA. The correlations of TCGA-annotated purity vs. the ABSOLUTE and InfiniumPurify estimates had been solely 0.16 and 0.10, respectively. These correlations had been decrease than our noticed correlation between TPF and purity (r~0.4).
Neural community classification of most cancers subtypes
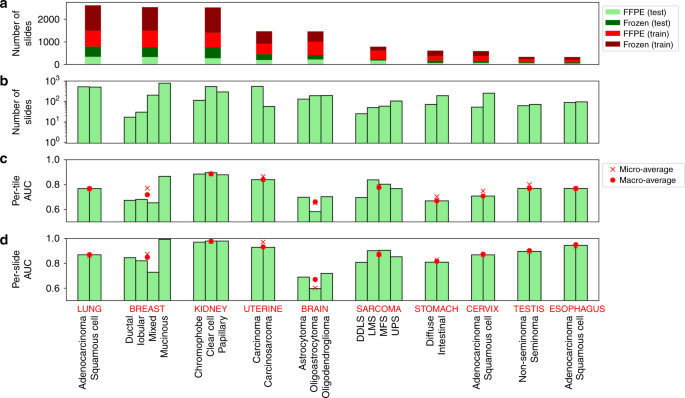
We additionally utilized our algorithm to categorise tumor slides primarily based on their most cancers subtypes (Fig. 1a). This evaluation was carried out on ten tissues for which pathologist subtype annotation was obtainable on TCGA: sarcoma (SARC), mind (LGG), breast (BRCA), cervix (CESC), esophagus (ESCA), kidney (KIRC/KIRP/KICH), lung (LUAD/LUSC), abdomen (STAD), uterine (UCS/UCEC), and testis (TGCT). Most cancers subtypes with at the very least 15 samples had been thought of, primarily based on TCGA metadata (see “Strategies”). As a result of comparable numbers of FFPE and flash-frozen samples are current in TCGA most cancers sorts (FFPE to frozen slide ratio: 1.0 ± 0.5), each had been included (Fig. 3a), and every tissue was stratified into its obtainable subtypes (Fig. 3b and “Strategies”). We used the identical CNN mannequin as for tumor/regular classification; nevertheless, for most cancers sorts with greater than two subtypes, a multi-class classification was used.
Determine 3c, d reveals the per-tile and per-slide classification outcomes (AUC ROCs alongside their micro- and macro-averages). On the slide degree, the classifiers can determine the subtypes with good accuracy in most tissues, although typically not but at medical precision (AUC micro-average: 0.87 ± 0.1; macro-average: 0.87 ± 0.09). The tissue with the very best AUC micro/macro-average was kidney (AUC 0.98), whereas the bottom was a mind with micro-average 0.60 and macro-average 0.67. All CIs had been above the 0.50 null AUC expectation, and all the AUCs had been statistically important (5% FDR, Benjamini–Hochberg correction32). For full CIs and P values, see Supplementary Information 1. The person subtype with the very best AUC is the mucinous subtype for breast most cancers (adjusted P worth <1e-300). The weakest P worth (adjusted P = 0.012) belongs to the oligoastrocytoma subtype of the mind. Slide predictions are superior to these on the tile degree, although with related traits throughout tissues. This means that tile averaging gives substantial enchancment of sign to noise, according to observations for the tumor/regular evaluation. In distinction to tumor/regular classification reaching excessive AUC’s throughout all most cancers sorts, subtype classification AUCs are decrease and span a wider vary. This implies that subtype classification is inherently more difficult than tumor/regular classification, with a narrower vary of picture phenotypes.
The pictures used within the subtype evaluation had been from a mix of frozen and FFPE samples. Though FFPE samples are most popular as a result of they keep away from distortions brought on by freezing, we examined whether or not the CNNs had been capable of classify subtypes for every pattern preparation (Supplementary Fig. 4). The CNNs categorized the FFPE and frozen samples with comparable accuracy, with the identical tumor sorts doing higher (e.g., kidney), or worse (mind) in every. Correlations between classification AUCs had been excessive throughout the 2 pattern preparations (r = 0.87 for macro-averages; r = 0.78 for micro-averages). As anticipated, FFPE-based classifications had been typically higher, notably for mind and sarcoma samples.
Cross-classifications between tumor sorts show conserved spatial behaviors
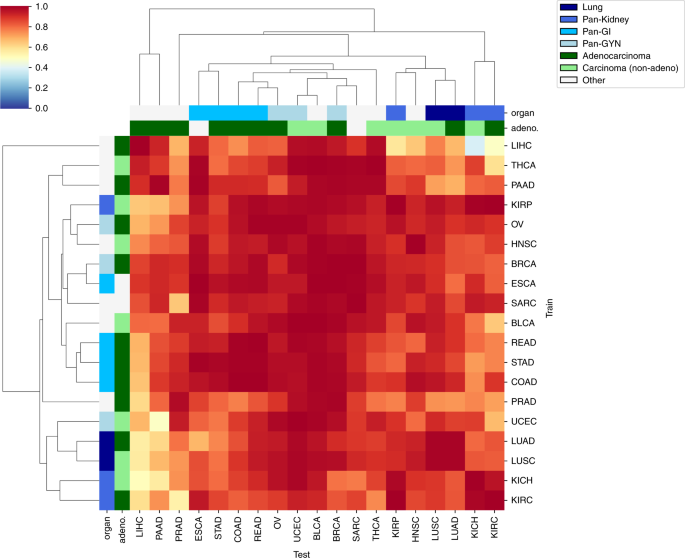
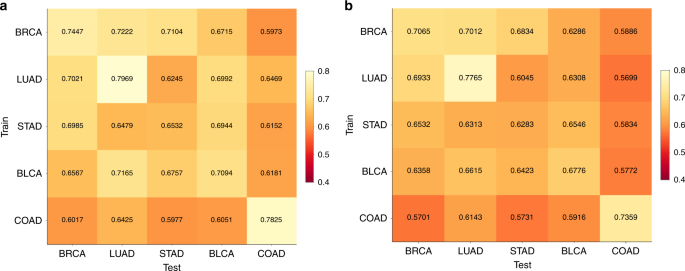
We subsequent used cross-classification to check the speculation that completely different tumor sorts share CNN-detectable morphological options distinct from these in regular tissues. For every most cancers kind, we re-trained the binary CNN classifier for tumor/regular standing utilizing all flash-frozen WSIs within the set. We then examined the flexibility of every classifier to foretell tumor/regular standing within the samples from one another most cancers kind. Determine 4 reveals a heatmap of per-slide AUC for all cross-classifications, hierarchically clustered on the rows and columns of the matrix. A non-clustered model is offered in Supplementary Fig. 5 with CIs. Surprisingly, neural networks educated on any single tissue had been profitable in classifying most cancers vs. regular in most different tissues (common pairwise AUCs of off-diagonal components: 0.88 ± 0.11 throughout all 342 cross-classifications). This prevalence of sturdy cross-classification helps the existence of morphological options shared throughout most cancers sorts however not regular tissues. Particularly, classifiers educated on most most cancers sorts efficiently predicted tumor/regular standing in BLCA (AUC = 0.98 ± 0.02), UCEC (AUC = 0.97 ± 0.03), and BRCA (AUC = 0.97 ± 0.04), suggesting that these cancers most clearly show options common throughout sorts. At a 5% FDR, 330 cross-classification AUCs are important (See Supplementary Fig. 5 for statistical particulars). The AUC imply and CI decrease certain are every above 80% for 300 and 164 of those cross-classifications, respectively. A number of most cancers sorts, e.g., LIHC and PAAD, confirmed poor cross-classification to different tumor sorts, suggesting morphology distinct from different cancers.
The hierarchically clustered heatmap reveals pairwise AUC values of CNNs educated on the tumor/regular standing of 1 most cancers kind (prepare axis) examined on the tumor/regular standing of one other most cancers kind (take a look at axis). Adeno-ness (adenocarcinoma vs. non-adenocarcinoma) and organ of origin (lung, kidney, gastrointestinal, gynecological) for every set are marked with colours on the margins. Cancers with ambiguous or blended phenotype are marked as “Different”.
To enhance spatial understanding of those relationships, we examined how nicely tile-level predictions are conserved between completely different classifiers (Fig. 5), whereas additionally analyzing the impact of various the take a look at set. For every pair of classifiers, we specified a take a look at set then computed the correlation coefficient of the anticipated tumor/regular state (logit of the tumor chance) throughout all tiles within the take a look at set. We repeated this calculation for every take a look at set, which we listed by tissue kind (breast, bladder, and many others.). Every take a look at set included each tumor and regular slides for the tissue kind. Determine 5a, b reveals for every pair of classifiers the common and most correlation coefficients, respectively, over take a look at units. Many correlations are optimistic, with a median and normal deviation over all pairs of classifiers of 45 ± 16% (Fig. 5a, diagonal components excluded), indicating cross-classifiers agree on the tile degree. These tile-level outcomes supported the slide-level outcomes. Classifiers with low cross-classification slide-level AUCs, corresponding to LIHC, had the smallest tile-level correlations. Tile predictions additionally confirmed similarities between classifiers derived from the identical tissue (e.g., LUAD-LUSC, KICH-KIRP-KIRC). Similarities between classifiers turned much more obvious once we targeted on the take a look at tissue with the strongest correlation for every pair of classifiers (Fig. 5b). These optimistic correlations will not be merely resulting from distinguishing tiles in tumor slides from tiles in regular slides. Determine 5c, d is analogous to Fig. 5a, b, however computed solely over tumor slides. The outcomes are practically unchanged, indicating that they replicate conduct inside tumor pictures.
Correlations of predicted tumor/regular standing (i.e., logit of tumor chance) between pairs of classifiers, specified on the x and y axis. Correlations are first calculated utilizing the tile values for all slides of a given take a look at tissue. a Common correlation throughout tissues, utilizing each tumor and regular slides within the tissue take a look at units. b Correlation for the tissue set with the maximal correlation, utilizing each tumor and regular slides within the tissue take a look at units. c Common correlation throughout tissues, utilizing solely tumor slides within the tissue take a look at units. d Correlation for the tissue set with the maximal correlation, utilizing solely tumor slides within the tissue take a look at units.
We hypothesized that sure tissue sorts is likely to be notably simple to categorise, and to check this we tabulated which tissue units yielded the maximal correlations for every pair of classifiers in Fig. 5b (Supplementary Information 2). For every pair, we listed the three tissue units yielding the very best correlations. If this had been random, we might count on every tissue to look on this record 27 occasions. Nonetheless, we noticed excessive prevalence for BRCA (132 appearances, P = 8.5e-119), BLCA (106 appearances, P = 2.5e-43), and UCEC (62 appearances, P = 1.9e-11). Many classifier pairs agree higher inside these three tissues than they do inside their coaching tissues. Thus BRCA, BLCA, and UCEC are canonical sorts for intraslide spatial evaluation, along with their excessive cross-classifiability on the whole-slide degree (Fig. 4).
We in contrast the impact of minor modification to the structure on tumor/regular self- and cross-classifications. If we simply used the Inception v3 structure with out the extra dense layers (see “Strategies”), the outcomes had been inferior (Supplementary Fig. 6). Our structure (Fig. 1a) achieved a barely increased AUC on common (0.04 ± 0.068) in comparison with the unique Inception V3 community (Wilcoxon signed-rank take a look at P worth <1e-20).
Cross-classification relationships recapitulate most cancers tissue biology
To check the organic significance of cross-classification relationships, we assessed associations between the tissue of origin22 and cross-classification clusters. Particularly, we labeled KIRC/KIRP/KICH as pan-kidney33, UCEC/BRCA/OV as pan-gynecological (pan-gyn)34, COAD/READ/STAD as pan-gastrointestinal (pan-GI)35, and LUAD/LUSC as lung. The hierarchical clustering in Fig. 4 reveals that cancers of comparable tissue of origin cluster nearer collectively. We noticed that the lung set clusters collectively on each axes, Pan-GI clusters on the take a look at and partially the prepare axis, and Pan-Gyn additionally partially clusters on the take a look at axis. Pan-Kidney partially clusters on each axes. To quantify this, we examined the associations between proximity of cancers on every axis and similarity of their phenotype (i.e., tissue of origin/adeno-ness). Organ of origin was considerably related to smaller distances within the hierarchical clustering (P worth = 0.002 for take a look at axis and P = 0.009 for prepare axis; Gamma index permutation take a look at, see “Strategies”). We additionally grouped cancers by adenocarcinoma/carcinoma standing (Fig. 4, second row from high). Since SARC doesn’t match both class, and ESCA incorporates a mix of each classes, these two cancers had been labeled as “different”. The inter-cancer distances had been considerably related to adeno-ness on the prepare axis (P worth = 0.015). We noticed different intriguing relationships amongst cross-tissue classifications as nicely. Significantly, Pan-GI created a cluster with Pan-Gyn, supporting these tumor sorts having shared options associated to malignancy. Likewise, Pan-Kidney and lung additionally cluster shut to one another.
Validation of cross-classification relationships utilizing CPTAC pictures
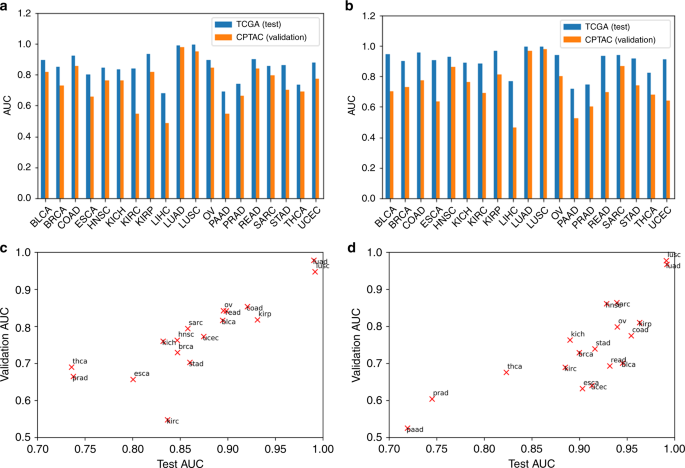
To validate the educated CNNs and their cross-classification accuracies, we utilized them to the LUAD and LUSC slides of the Scientific Proteomic Tumor Evaluation Consortium (CPTAC) dataset (see “Strategies”). TCGA-trained LUAD and LUSC classifiers had been extremely efficient on the CPTAC LUAD and LUSC datasets (Fig. 6a, b). The TCGA-trained LUAD and LUSC classifiers have validation AUCs of 0.97 and 0.95, respectively, on the CPTAC-LUAD dataset, and have validation AUCs of 0.97 and 0.98, respectively, on the CPTAC-LUSC dataset. Each of the TCGA-trained CNNs yielded well-separated distributions of TPF between CPTAC tumor and regular slides (Supplementary Fig. 7). CNNs educated on different TCGA tissue sorts had been additionally comparatively efficient on the CPTAC units, with common AUC of 0.75 and 0.73 when utilized to the CPTAC-LUAD and LUSC picture units, respectively. This was decrease than the efficiency of the TCGA-trained classifiers on the TCGA LUAD and LUSC units (common AUC 0.85 and 0.90, respectively), suggesting that cross-classification is extra delicate to batch protocol variations. Nonetheless, the correlation between AUCs on the TCGA and CPTAC units was excessive (Fig. 6c, d: LUAD: r = 0.90, LUSC: r = 0.83), indicating that relationships between tumor sorts have a transparent sign regardless of such sensitivities. The CPTAC-LUAD and LUSC datasets had been additionally used to coach classifiers, which had been then examined on the TCGA most cancers units. We noticed excessive correlation between TCGA-trained and CPTAC-trained cross-classification AUCs (Supplementary Fig. 8, LUAD: r = 0.98, LUSC: r = 0.90).
Classification AUCs of every TCGA-trained tumor/regular classifier utilized to LUAD and LUSC pictures from TCGA (reserved “take a look at” information) and CPTAC (exterior “validation” information, LUAD: n = 1055, LUSC: n = 1060) are proven. a Bar graphs evaluating take a look at and validation AUCs on LUAD and b LUSC slides. c Scatter plot of take a look at AUC versus validation AUC for LUAD and d LUSC. TCGA prepare and take a look at pattern sizes are offered in Fig. 2a.
Comparisons of neural networks for TP53 mutation classification
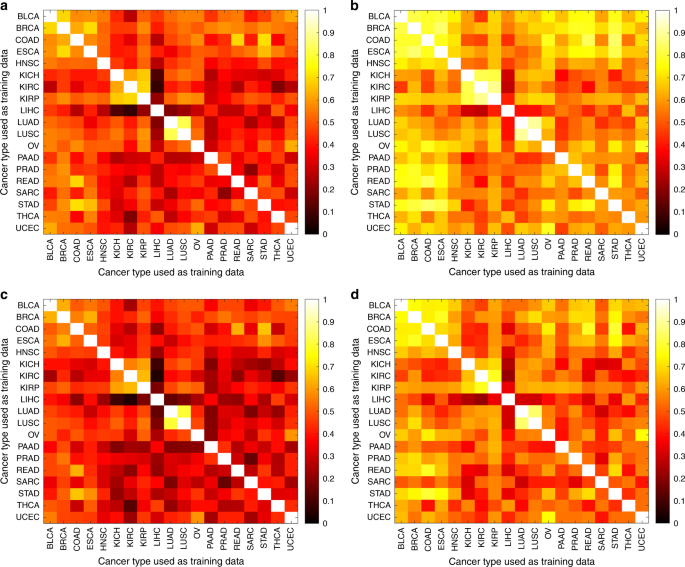
To analyze how pictures can be utilized to tell apart most cancers drivers, we examined the accuracy of CNNs for classifying TP53 mutation standing in 5 TCGA most cancers sorts, particularly BRCA, LUAD, STAD, COAD, and BLCA. We selected these resulting from their excessive TP53 mutation frequency36,37,38, offering adequate testing and coaching units for cross-classification evaluation. Utilizing switch studying, we obtained reasonable to low AUCs for TP53mut/wt classification (0.66 for BRCA, 0.64 for LUAD, 0.56 for STAD, 0.56 for COAD, and 0.61 for BLCA). Because of this weak efficiency, we switched to a extra computationally intensive method by which we totally educated all parameters of the neural networks primarily based on an structure described in ref. 19 (Fig. 1b), with undersampling to handle information imbalance and a 70/30 ratio of slides for coaching and testing. Determine 7a, b reveals heatmaps of AUC for the per-tile and per-slide classification outcomes, respectively (see additionally Supplementary Figs. 9 and 10). Self-cohort predictions (diagonal values) have AUC values starting from 0.65–0.80 for per-slide and 0.63–0.78 for per-tile evaluations. Abdomen adenocarcinoma (slide AUC = 0.65) was notably tougher to foretell than lung adenocarcinoma (slide AUC = 0.80), for which we discovered AUC values similar to the AUC = 0.76 LUAD outcomes reported by Coudray et al.19. This LUAD totally educated community (AUC = 0.76) outperformed the switch studying for a similar information (AUC = 0.64). The CNNs achieved the next AUC in contrast with a random forest utilizing tumor purity and stage for TP53 mutation prediction (see Supplementary Fig. 11), suggesting the CNNs use extra refined morphological options of their predictions. We additionally noticed that CNNs had been capable of extra precisely determine tumors with TP53 mutations when the allele frequency of the mutation was increased, suggesting that prediction is less complicated when the tumor is extra homogeneous (Supplementary Fig. 12). The F1 scores of the CNNs are offered in Supplementary Fig. 13.
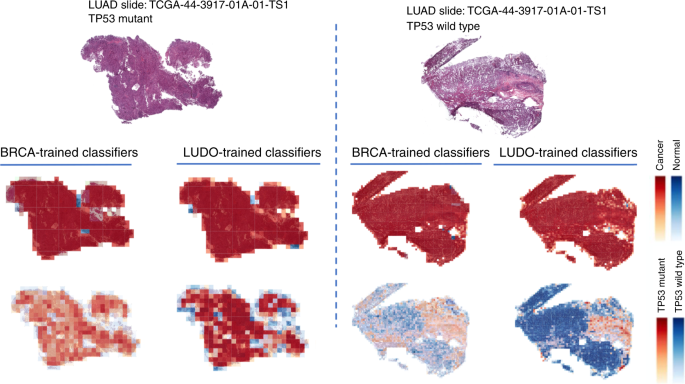
We additionally examined the flexibility of the TP53 CNNs to cross-predict throughout most cancers sorts. Cross-predictions yielded AUC values with a comparable vary because the self-cohort analyses (AUCs 0.62–0.72 for slides; 0.60–0.70 for tiles), although self-cohort analyses had been barely extra correct. These AUC values will not be adequate for sensible use, although the optimistic cross-classification outcomes recommend that it is likely to be attainable to mix datasets to extend accuracy (see “Dialogue”). Colon adenocarcinoma AUC values tended to be low as each a take a look at and prepare set, suggesting TP53 creates a unique morphology on this tissue kind. Total, the optimistic cross-classifiabilities assist the existence of shared TP53 morphological options throughout tissues. Determine 8 reveals TP53 mutational heatmaps of 1 LUAD slide recognized to be mutant and one LUAD slide recognized to be wild kind from the sequencing information. We in contrast the LUAD- and BRCA-trained deep studying fashions on these slides, as these two fashions offered the very best AUC values in our cross-classification experiments. Prediction maps for tumor/regular standing (second row) and TP53 mutational standing (third row) are proven for each samples. Each tumor/regular fashions accurately predicted the vast majority of tiles in every pattern as most cancers. Analogously, the BRCA-trained TP53 mutation standing mannequin predicts patterns much like the LUAD-trained mannequin. Importantly, the tumor/regular and TP53mut/wt classifiers spotlight completely different areas, indicating these classifiers are using distinct spatial options. A caveat to those analyses, nevertheless, is that the spatial variation inside heatmaps could replicate TP53mut-associated microenvironmental options fairly than genetic variation amongst most cancers cells.
The primary row reveals two LUAD H&E slides with TP53 mutant (left panel) and wild kind (proper panel). The second row reveals prediction maps for these two slides utilizing tumor/regular classifiers educated on BRCA and LUAD samples. Each fashions efficiently classify samples as most cancers and predict related heatmaps. The third row reveals prediction maps for these slides utilizing TP53 mutation classifiers educated on BRCA and LUAD. The BRCA-trained and LUAD-trained heatmaps are related, suggesting that there are spatial options for TP53 mutation which can be sturdy throughout tumor sorts.
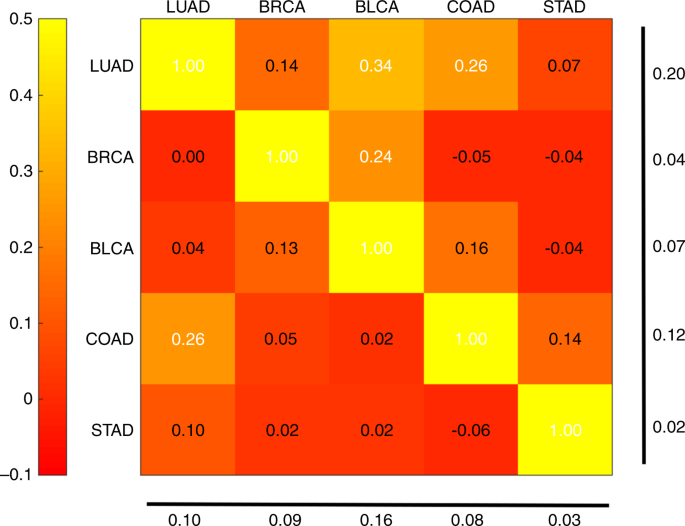
We subsequent carried out a tile-level cross-classification evaluation as a operate of take a look at set. For many take a look at most cancers sorts, we noticed little correlation when evaluating networks educated on cancers “A” and “B” utilized to check most cancers “C”. Subsequently, we targeted on circumstances the place C is similar as B. Determine 9 plots the correlations of TP53 mutation chance logits throughout most cancers pairs, the place every row denotes the most cancers kind the primary CNN is educated on, and every column is each the take a look at tissue and the second CNN coaching tissue. In these circumstances, the correlation coefficients had been typically optimistic and met statistical significance although with reasonable magnitude. All correlations had been important, apart from the BRCA TP53 classifier utilized to LUAD tumors (t take a look at on Fisher z-transformed correlation coefficients, FDR 5%). Notably, classifiers primarily based on LUAD, BRCA, and COAD labored nicely on BLCA, BLCA, and COAD tumors, respectively. BLCA and LUAD are the 2 take a look at cancers with the biggest correlations (column common). LUAD and COAD are the 2 coaching cancers with the biggest correlations (row common). The excessive row and column averages for LUAD point out it’s canonical each as a take a look at and a coaching set. Apparently, the correlations of Fig. 9 will not be symmetric. For instance, the community educated on LUAD achieves a correlation of 0.34 on BLCA, whereas the community educated on BLCA has a correlation of 0.04 when examined on LUAD.
Row labels denote the most cancers kind used to coach the primary TP53 mutation classifier. Column labels denote each the take a look at tissue and the tissue of the second TP53 classifier. Heatmap values point out correlation coefficients of mutation chance logits between the 2 classifiers on the take a look at tissue. Numbers on the backside and proper present column and row averages, respectively, with diagonal values excluded.
Options impacting tumor purity prediction
TCGA gives annotations solely on the whole-slide degree, limiting our capability to construct classifiers that resolve predictive options. To higher examine options, we obtained datasets with increased decision annotations, i.e., BreCaHAD39 which gives nucleus-level tumor/regular annotations of 162 breast most cancers ROIs (see the strategies part for particulars), and eight colorectal ROIs hand-annotated at nuclear decision (>18,000 cells) by our group. These annotations present the bottom fact tumor purity (the fraction of tumor cells, aka mobile malignancy) used for the evaluation. We then analyzed these information on the tile degree (512 × 512 pixels). We educated CNNs vs. the bottom fact purity values for every tile, randomly splitting the BreCaHAD dataset into 150 prepare and 12 take a look at ROIs (a complete of >23,000 cells) and utilizing the colorectal set for validation. Purities of the colorectal tiles are unfold over a variety (imply 58%, normal deviation 19.2%), whereas BreCaHAD purities are increased (imply 87% within the coaching set), as detailed in Supplementary Fig. 14. These CNNs yielded a imply absolute error of 14% and 15% for the take a look at breast and colorectal units, respectively. Root imply squared error (RMSE) values had been 8% and 20%, respectively. The common prediction for the colorectal datasets (69%) was intermediate between the true colorectal imply (58%) and the breast imply (87%), suggesting that, though the CNN was educated solely on breast information, the CNN was capable of be taught some options widespread between breast and colorectal tumors. As a comparability, we additionally calculated RMSEs between purity and TPF as predicted by the TCGA-trained BRCA and COAD classifiers on the colorectal set. These RMSEs had been 45% and 39%, respectively. These values had been inferior to the BreCaHAD-trained CNNs, indicating that nuclear annotations present extra predictive data past the general slide label.
We additional examined whether or not purity was being predicted from solely the picture areas containing particular person nuclei, or whether or not intercellular data was getting used. For this, we made use of a CNN classifier40 that predicts tumor/regular standing from particular person nucleus pictures (see “Strategies”). We educated on the breast nuclei, and this was capable of predict tumor standing of reserved breast nuclei pictures with excessive accuracy (AUC 93–98%) Nonetheless, the breast-trained CNN yielded poor predictions on the colorectal nuclei (AUC 56%). We examined whether or not including up the contributions of nuclei inside every ROI would result in good predictions on the ROI degree. Nonetheless, the common RMSE throughout colorectal ROIs was 25%, increased than the RMSE from the tile-based evaluation (20%) of the identical information. This implies that, though the tile-based method just isn’t conscious of particular person cells, it compensates through the use of intercellular areas of pictures.