This part presents the experimental outcomes of the utilized evaluation. The computational outcomes are adopted by experimental validations.
Gene co-expression evaluation
The coaching dataset was analyzed utilizing the WGCNA methodology to create a co-expression community and establish gene modules that have been extremely correlated with given phenotypes. WGCNA makes use of the scale-free topology criterion to assemble a gene co-expression community17. The selection of the gentle thresholding energy (β) is essential because it determines the co-expression similarity to calculate adjacency. To establish the most effective gentle threshold, a community topology evaluation was performed for varied values, as proven in Supplementary Fig. S1. The dimensions-free topology match index curve’s highest worth was obtained earlier than it flattened out, yielding the gentle threshold worth of “8”.
Determine 1a signifies the correlation coefficient and p-value that characterize the connection between the respective module eigengenes (in rows) and pattern phenotype (in columns) for the coaching set. A dynamic tree-cutting approach was utilized to establish modules which have comparable gene expression profiles18. On this regard, a threshold of 0.25 peak reduce was set which corresponds to a correlation of 0.75. The modules with comparable expression profiles have been mixed primarily based on this threshold, leading to 22 totally different modules. After analyzing the correlation and p-values, we chosen six vital modules: the m2 module is for the liver metastasis from the first CRC phenotype; m6 and m7 characterize the first CRC phenotype; m16, m17, and m18 are vital modules for the liver metastasis from the traditional colon tissue phenotype.
Identification of essentially the most associated co-expression modules (row) to particular phenotypes (column). The scale of every circle signifies the p-value, whereas the colour represents the Pearson correlation. (a) The correlation heatmap for the coaching dataset. (b) The correlation heatmap for the validation dataset. The unique heatmaps of the WGCNA methodology are reorganized utilizing the ggpubr R-library19.
The numerous modules have been merged to characterize associated genes right into a important module for every phenotype. The members of those important modules present reverse mRNA expression patterns, e.g. the module members are down-regulated in regular colon samples, identical genes are up-regulated in major CRC samples. Supplementary Desk S1 exhibits the variety of genes in the principle module of every phenotype. An enormous variety of genes (#7537) have been noticed within the “major from regular” module, then adopted by “metastasis from regular” (#2453) and “metastasis from major” (#733) modules.
Community clustering and submodule choice
The tissue-specific interplay networks of the chosen three phenotypes have been constructed primarily based on associated tissue. For the “metastasis from major” module, its useful interplay community (FIN) was constructed on the liver tissue-specific community; for the “major from regular” module, the FIN was constructed on the colon tissue-specific FIN; for the “metastasis from regular” module, the FIN was constructed on the liver tissue-specific FIN. The whole variety of genes and interactions in these FINs are given in Supplementary Desk S2. Then network-based clustering algorithms run on these FINs. The efficiency of every clustering algorithm was calculated with analysis metrics. The obtained outcomes for the three phenotypes are summarized within the following sections.
Submodules for metastasis from major colon samples
Markov clustering (MCL), fuzzy neighborhood (FN), spectral clustering algorithms in addition to Infomap and Label Propagation (LP) have been used within the FIN created for this phenotype. 5 clustering algorithms run on the identical FIN. The efficiency of every algorithm was evaluated utilizing each inside and organic metrics, these evaluations are summarized in Supplementary Fig. S2. Contemplating analysis metrics, the LP and Infomap algorithms achieved the most effective clustering outcomes. The submodules detected by these two algorithms have been re-evaluated with their particular person Organic Homogeneity Index (BHI), Wang Organic Course of (Wang-BP), and Molecular Perform (Wang-MF) metrics. First, submodules with the very best BHI, Wang-BP, and Wang-MF values have been analyzed, moreover, the presence of considerably regulated genes within the related submodule was thought of. Accordingly, the evaluation was utilized to submodules given in Supplementary Desk S3 for biomarker choice.
Because of these analyses, the submodules 1, 12, and 13 of the Infomap algorithm and the submodule 2 of the LP algorithm have been decided for additional biomarker evaluation. When all these submodules are thought of, 57 genes represented elevated expression values in metastatic samples in comparison with the first CRC.
Submodules for major from regular samples
All clustering algorithms have been used within the FIN constructed for the phenotype of major CRC developed from regular colon samples (Supplementary Fig. S3). FN and Spectral algorithms led to the most effective outcomes. The submodules detected by these two algorithms have been re-evaluated by metrics as given in Supplementary Desk S4. Consequently, submodules 2 and seven within the FN algorithm and submodules 2 and 16 within the Spectral algorithm have been chosen for additional evaluation. Inside totally different submodules, 120 genes have elevated expressions in major CRC samples in comparison with the traditional colon group. However, 9 genes confirmed decreased expressions in the identical affected person teams.
Submodules for metastasis from regular samples
5 clustering algorithms have been utilized to establish biomarker genes that play an necessary position in liver metastasis improvement from regular colon samples (Supplementary Fig. S4). We noticed that LP and Infomap algorithms created too many clusters having few members. Because the outcomes of the FN algorithm are fairly constant, essentially the most vital submodules fashioned by this algorithm have been chosen for additional evaluation. For this course of, the submodules detected by the FN algorithm have been re-evaluated by organic metrics summarized in Supplementary Desk S5; the presence of genes that modified considerably within the related submodule was additionally thought of. Consequently, submodules 1, 4, 5, and 12 have been discovered to be vital. When these submodules have been evaluated, 107 and 10 genes confirmed elevated and decreased expressions, respectively in metastatic samples in comparison with the traditional colon group. Supplementary Desk S6 lists the genes in all key submodules recognized by the clustering algorithms within the related phenotype.
Validation set evaluation
Statistically vital modules have been additionally decided by making use of WGCNA to the validation dataset. The Pearson correlation and p-values of the chosen modules are given in Fig. 1b.
4 vital modules have been extracted. Two of modules have been within the “major colon developed from regular colon tissue” phenotype (m5, m12), and two of them have been within the “major colon and metastasis developed from regular colon tissue” phenotype (m14, m15). Members of modules related to the identical phenotype have been mixed and two large modules have been obtained, the knowledge on these modules is given in Supplementary Desk S7.
Based mostly on related phenotypes, the genes within the vital modules recognized by WGCNA on the validation and coaching datasets have been in contrast, and customary genes throughout the identical phenotypes in each datasets have been extracted. Supplementary Desk S8 summarizes the variety of these widespread genes. Supplementary Desk S9 lists their names, gene expression ranges, and associated phenotypes.
Biomarker choice
A gene enrichment evaluation was carried out for widespread genes given in Supplementary Desk S9. Closing biomarkers have been chosen from genes with elevated or decreased mRNA expression profiles and concerned in vital organic processes or pathways.
Biomarkers for major CRC
Considerably expressed genes for the event of major CRC from regular colon tissue have been explored. Elevated ranges of mRNA expression have been noticed for all genes. Based mostly on the gene set enrichment evaluation, vital KEGG pathways embrace the cell cycle, p53 signaling pathway, and mobile senescence. Vital organic processes cowl constructive regulation of the cell cycle course of, regulation of G2/M transition of the mitotic cell cycle, G1/S transition of the mitotic cell cycle, regulation of sign transduction by p53 class mediator, regulation of cell inhabitants proliferation, and regulation of the apoptotic course of. Particulars relating to the gene set enrichment evaluation for major CRC are offered in Supplementary Desk S10.
Biomarkers for major CRC and metastasis improvement
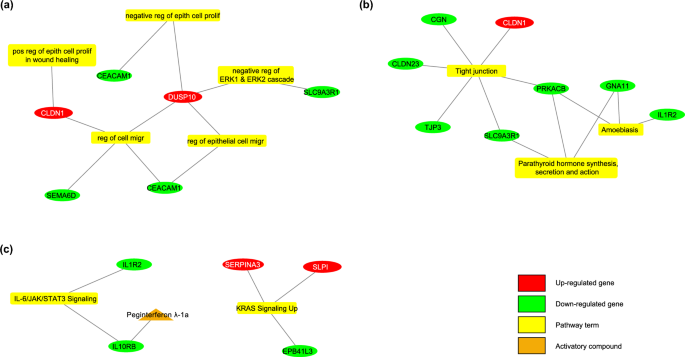
Genes which can be considerably expressed within the improvement of major CRC and metastasis from regular colon tissue have been examined. Based mostly on the gene set enrichment evaluation, necessary organic processes embrace damaging regulation of ERK1 and ERK2 cascade and regulation of cell migration. Vital hallmarks of most cancers phrases embrace KRAS signaling up and IL-6/JAK/STAT3 signaling. DUSP10, CLDN1, SERPINA3, and SLPI genes, that are concerned in necessary organic processes, present elevated expression in each datasets. It was decided that SLC9A3R1, CEACAM1, IL10RB, and IL1R2 genes confirmed decreased expression in each coaching and validation datasets for major CRC and metastasis. Particulars relating to the gene set enrichment evaluation for major CRC and metastasis are offered in Supplementary Desk S11.
Therapeutic medicine
The biomarkers summarized Supplementary Desk S12 have been mutually noticed in vital modules for each coaching and validation datasets, thus they have been offered as enter to seek for medicine that may therapeutically goal these proteins. The medicine, that may therapeutically goal the given biomarkers, have been searched from the Drug-Gene Interplay Database (DGIdb)20 as defined within the methodology part.
Medicine prompt for treating major CRC
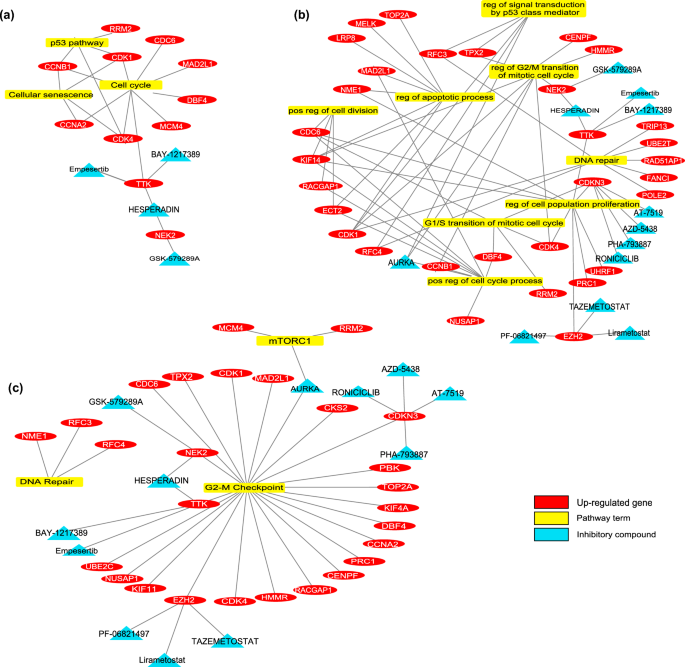
There are 61 biomarker genes with elevated expressions within the regular colon and first CRC samples. Because of drug screening, many medicine have been retrieved to focus on these proteins. Determine 2 exhibits the enrichment outcomes of goal biomarkers, related pathways / processes and their therapeutic medicine. The massive drug listing was re-scanned within the literature through the use of “tumor development”, “proliferation”, “cell dying”, “apoptosis”, “autophagy”, “G1-S arrest”, “invasion”, “EMT mechanism”, or “colorectal most cancers” key phrases. Because of all these analyses, extra particular protein targets and medicines have been recognized. Supplementary Desk S12 exhibits the restricted drug listing. Amongst these medicine, the BAY-1217389 and Hesperadin compounds, which inhibit TTK protein kinase, have been thought of for in-vitro experiments to indicate their efficacy within the major CRC cell traces.
Medicine prompt for treating liver metastatic CRC
Within the pattern group of regular colon, major CRC, and liver metastasis, there have been 42 genes (34 of them with decreased, 8 of them with elevated mRNA expression). Determine 3 exhibits the enrichment outcomes of biomarkers and their therapeutic medicine. The medicine focusing on biomarkers, which work in cancerization and metastasis processes, have been investigated. There was no inhibitory group that targets proteins with elevated expression ranges. Among the many biomarkers with decreased expressions, the “Peginterferon λ-1a” from the activator group was detected for focusing on IL10RB, which is a member of the IL-6/JAK/STAT3 signaling pathway (Supplementary Desk S13). This agent was used to show its in-vitro efficacy within the liver metastatic CRC cell line.
Experimental validation
Hesperadin decreases cell viability in short-term cytotoxic impact
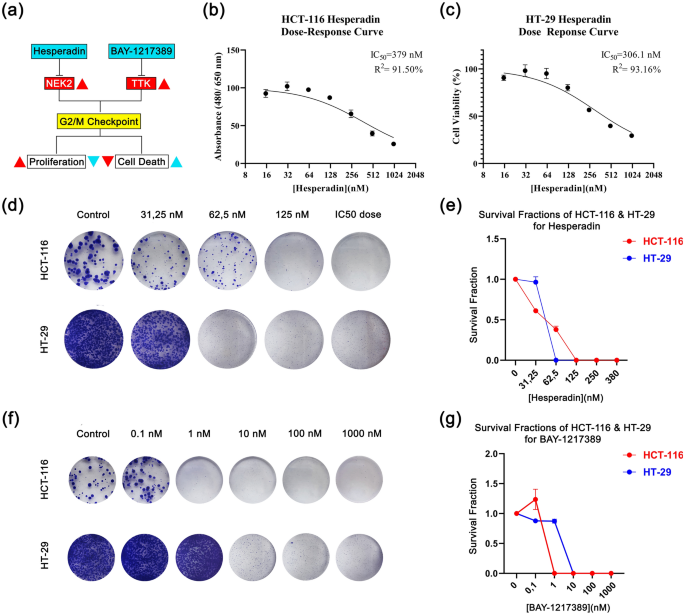
In-silico evaluation predicts that TTK and NEK2 are promising targets to deal with the proliferative habits of CRC. The community evaluation signifies these proteins are associated to cell division and proliferation. Due to this fact, the in-silico mannequin means that their potent inhibitors Hesperadin and BAY-1217389 (BAY-12) might decline most cancers development (Fig. 4a).
Cell viability and survival graphs in response to drug therapies. (a) A primary graphical summary of within the vitro pipeline indicating hesperadin and BAY-1217389’s mechanism of anti-proliferative and cytotoxic results is proven. Whereas crimson shade represents most cancers standing, gentle blue serves as therapy standing. The route of the arrows factors out a rise (▲) and a lower (▼). (b) Hesperidin dose–response curve in HCT-116. (c) Hesperadin dose response curves in HT-29. Colony photographs together with relative dose/survival fraction graphs are proven for HCT-116 (d,e) and HT-29 (f,g).
First, we investigated if Hesperadin and BAY-12 might lower cell viability of grade II-III human colon carcinoma traces, HT-29 and HCT-116. The cells have been handled with 0.1–1000 nM concentrations of those brokers and cell viability was measured by formazone forming WST-1 assay. The outcome factors out that Hesperadin has a potent impact on the viability. Its IC50 values are 379 nM and 306.1 nM for HCT-116 and HT-29 respectively (Fig. 4b,c). Whereas Hesperadin abates cell viability of each traces inside this dosage vary in 48 h, BAY-12 signifies no cytotoxic impact. Nonetheless, we hypothesized that BAY-12 might have a cytostatic impact in the long run on account of its doable relation with the cell cycle.
BAY-1217389 led to a decline in colony formation in 14-days (long-term interval)
We carried out a colony formation assay to estimate if there’s a long run cytostatic impact on the cell reproductive system. Regardless that BAY-12 didn’t trigger a cytotoxic impact in 48 h within the WST-1 viability assay, it decreased the colony forming capability. In response to 10 nM of this agent, there was no colony counted after 14 days. Equally, Hesperadin inhibited clonogenic habits in a dose-dependent method. HCT-116 cells tolerated this agent higher than HT-29, according to their IC50 values. The survival fraction declined progressively at 15 to 62.5 nM focus and the colonies cleared away at 125 nM for HCT-116 (Fig. 4d,e). There have been no colonies noticed in response to a 62.5 nM dose of hesperidin for HT-29 (Fig. 4d,e). Thus, we recommend whereas HT-29 is extra susceptible to Hesperadin therapy, BAY-12 impacts each cells at decrease doses (10 nM) (Fig. 4f,g).
Regardless that each Hesperadin and BAY-12 have cytotoxic or cytostatic results on cell viability in CRC, BAY-12 was extra possible cytostatic in long-term intervals contemplating low response to WST-1 whereas excessive efficiency in colony formation. Each candidates have anti-proliferative results on CRC.
Hesperadin and BAY-1217328 have anti-proliferative impact
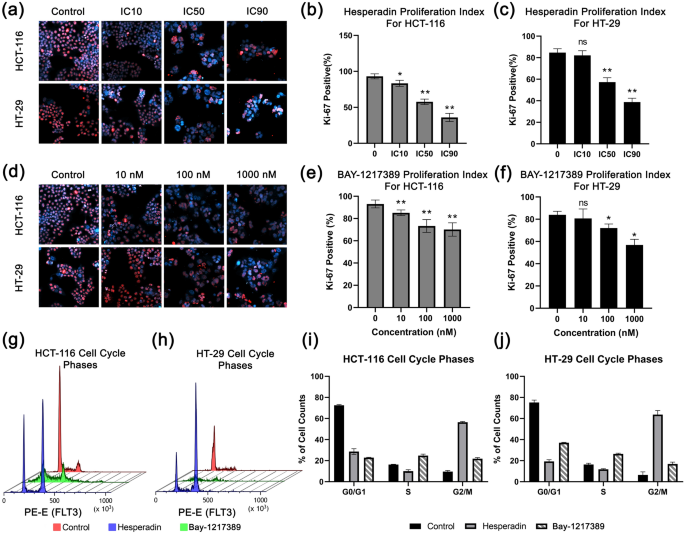
The impact on viability was researched whether or not it was an anti-proliferative or cytotoxic response. Immunofluorescence Ki-67 stainings have been carried out to judge proliferative cells. The proliferation index of each traces was decreased in a dose dependent response in opposition to Hesperadin. Whereas the Ki-67 constructive cells have been 80% for management samples, it was 57.25% and 52.68% for IC50 concentrations of this agent in HT-29 and HCT-116 cells, respectively (Fig. 5a–c). Though Hesperadin exhibits a barely stronger cytotoxic impact on HT-29 in account of WST-1 and colony formation assays, there isn’t any statistical distinction in Ki-67 rely.
Proliferation and cell cycle graphs. Immunofluoresense photographs of Ki-67 (crimson) as proliferation marker and Hoescht-33342 as counter staining (blue) in response to (a) hesperadin and (d) BAY-1217328 therapies are introduced. Bar graphs characterize proliferation indexes for every doses in accordance with Ki-67 constructive cell rely proportion in hesperadin handled for (b) HCT-116, for (c) HT-29 and BAY-1217389 handled teams for (e) HCT-116, for (f) HT-29. Cell cycle histograms are stand for (g) HCT-116 and (h) HT-29. Purple signifies management, blue is hesperadin handled and inexperienced is BAY-1217389 handled cells. Bar graphs characterize cell cycle phases percentages of cell counts are proven for (i) hesperadin and (j) BAY-1217328 therapies. “n.s” stands for non-significant. Asterisk (*) signifies p-value < 0.05, and double asterisk (**) means p-value < 0.01.
Whereas there have been no IC50 values measured by means of WST-1 viability assay in 48 h for BAY-12, 10 nM, or increased doses abated colony formation. Henceforward, the anti-proliferative impact of this agent was investigated in a variety of 10–1000 nM doses. The proliferation indexes of management samples have been 83.91% and 93.06%. This ratio was lowered to 72.03% and 73.30% in response to 100 nM of BAY-12 handled samples for HT-29 and HCT-116 cells, respectively (Fig. 5d–f). At increased doses, HT-29 was extra delicate to BAY-12. 1000 nM most dose decreased Ki-67 constructive cell to 56.90% for HT-29 whereas this highest focus diminishes the sign not more than 70.13% for HCT-116.
Hesperadin and BAY-1217389 arrest cell cycle on the G2/M section
As a result of the Ki-67 biomarker signifies that Hesperadin and BAY-12 have a level of anti-proliferative impact on cell viability and the recognized therapeutics goal cell cycle regulators TTK and NEK2, we examine through which section they hinder the cell cycle. To characterize the anti-proliferative mechanism of those medicine, the DNA quantity of pre-treated cells stained by Rnase I/ Propidium Iodide (PI) assay after 48 h incubation and measured in movement cytometry. As proven in Fig. 5g–j, these candidates arrest cell cycles on the G2/M section in HCT-116 and HT-29 cell traces. Hesperadin has a stronger impact on this habits (56.9%, 61.0% for HCT-116 and HT-29, respectively) than Bay-12 (22.6%, 18.1% for HCT-116 and HT-29, respectively).
IL-29A inhibits the metastatic habits in colorectal carcinoma
In-silico evaluation additionally predicts the agent Peginterferonλ-1A for metastatic CRC. Community evaluation prompt that the lower within the expression stage of IL10RB within the IL-6/JAK/STAT3 pathway in metastatic CRC sufferers could also be related to the metastatic phenotype. We thus verified the info derived from in-silico evaluation below in-vitro circumstances.
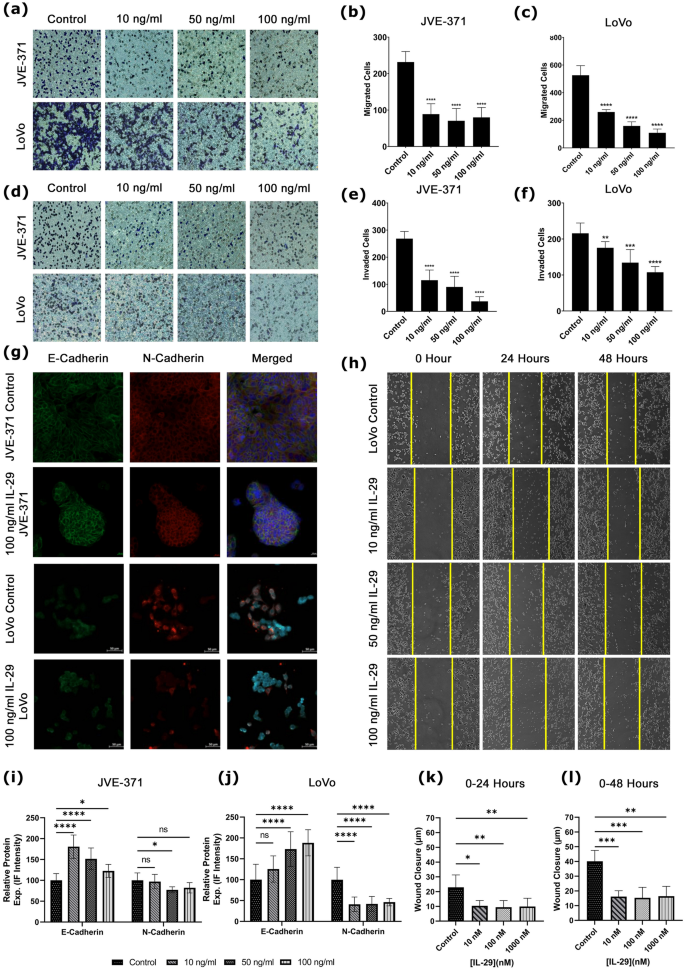
To validate the impact of the agent Peginterferonλ-1A for metastatic CRC, we investigated its non-pegylated kind, interferonλ-1A (IL-29A). Firstly, the effectivity of IL-29A on migration and invasion capacities of LoVo and JVE-371 metastatic CRC cell traces was recognized by transwell assay. IL-29A was handled with 10, 50, and 100 ng/ml concentrations on LoVo and JVE-371 cell traces at 48 h. Within the lymph node metastatic CRC LoVo cell line, IL-29A decreased the migration fee at 10, 50 and 100 ng/ml concentrations, respectively, in comparison with the TGFβ-induced management group (Fig. 6a–c, p-value < 0.0001). IL-29A additionally decreased invasion fee in comparison with the TGFβ-induced management group in growing concentrations on the LoVo cells (Fig. 6d,f, p-value = 0.0065, p-value = 0.0003 and p-value < 0.0001). Within the liver metastatic colorectal carcinoma JVE-371 cell line, migration, and invasion charges have been decreased in contrast with the TGFβ-induced management group (Fig. 6d,e p-value < 0.0001).
Migration, invasion and EMT capability in response to IL-29. (a) Cells migrated and (d) invaded the decrease membrane in transwell assay handled with IL-29 doses and management teams stained with crystal violet (darkish blue). Bar graphs characterize migrated cells by doses of IL-29 for (b) JVE-371 and (c) LoVo. Equally, invaded cells are introduced for (e) JVE-371 and (f) LoVo. (g) IL-29 metastatic cell traces are stained with EMT markers. Inexperienced stands for E-Cadherin, Purple is N-Cadherin, and Blue is Hoescht 33,342 (as counterstain). Relative protein expression is analyzed in accordance with IF depth and proven in bar graphs for (i) JVE-371 and (j) LoVo. (h) Wound therapeutic photographs are introduced. Wound closure in time (okay) 0–24 h and (l) 0–48 h are proven. Additionally, Bar graphs characterize cell cycle phases percentages of cell counts are proven for hesperadin (i) and BAY-1217389 (j) therapies. “n.s” stands for non-significant. Asterisk (*) signifies p < 0.05, double asterisks (**) means p-value < 0.01, Triple asterisks (***) means p-value < 0.001, and quadruple asteriks (****) signifies p-value < 0.0001.
Afterwards, epithelial-mesenchymal transition (EMT) was evaluated, since EMT is a extremely dynamic course of that enables cells to transition from the epithelial kind to the mesenchymal kind. Due to this fact, EMT results in the initiation of the metastatic cascade21. TGF-β induced JVE-371 cells to induce EMT confirmed a major lower within the quantity of E-cadherin (Fig. 6g, p-value < 0.0001) and a major improve within the quantity of N-cadherin (Fig. 6g,i p-value < 0.0001) in contrast with the non-TGF-β induced group. A major improve within the quantity of E-cadherin was noticed when IL29A was utilized to TGF-β-induced JVE-371 cells (Fig. 6g,i p-value < 0.0001, p-value = 0.0041). On the identical time, a major lower was noticed within the quantity of N-cadherin at 50 and 100 ng/ml concentrations (Fig. 6g,i, p-value < 0.0003, p-value < 0.0274), with none change at 10 ng/ml focus (Fig. 6i. p-value = 0.5951). In line with the immunofluorescence staining outcomes, no statistically vital change was noticed in LoVo cells at 10 ng/ml IL29A focus in comparison with the TGFβ-induced management group. Nonetheless, a statistically vital improve within the quantity of E-cadherin was noticed at 50 ng/ml and 100 ng/ml concentrations (Fig. 6g,j, P = 0.4706, p-value = 0.0012, and p-value < 0.0001). Likewise, a major lower was noticed within the quantity of N-cadherin, an indicator of mesenchymal phenotype, at growing concentrations of IL29A in LoVo cells in comparison with the TGFβ-induced management group (Fig. 6g,j, p-value < 0.0001).
To research migrative phenotype, a wound therapeutic assay was established along with a transwell migration assay. We monitored wound closure distance (µm) of LoVo cells for 48 h in management and IL-29 therapy teams. IL-29 therapy decreased the capability of wound closure of LoVo cells at 24 and 48 h, indicating that this therapy decreased migrative potential (Fig. 6h,okay,l). The info is correlated with the outcomes of the transwell migration assay. JVE-371’s development sample isn’t appropriate for wound therapeutic assay, subsequently we couldn’t monitor it on this experiment.
Total, we investigated the metastatic phenotype by transwell migration invasion capability, wound closure potential, and mesenchymal phenotype. The outcomes point out that in silico predicted IL-29A attenuates metastatic phenotypes in lymph metastatic LoVo and liver metastatic JVE-371 cells.