We recruited 460 CRC sufferers from a single hospital in Guangzhou (Strategies). All sufferers had been remedy naïve by the point of enrollment. Our cohort included sufferers with a large age vary, from 21 to 88 years outdated (Fig. 1a), with 95 sufferers recognized beneath the age of 40 and 167 sufferers beneath the age of fifty. Throughout all age teams (Supplementary Knowledge 1), there have been extra male sufferers than feminine sufferers. 14.8% (n = 68) of cancers had been stage I, 32.0% (n = 147) stage II, 36.1% (n = 166) stage III and 17.2% (n = 79) stage IV; 24.8% (n = 114) had been from the suitable hemicolon, 34.8% (n = 160) left hemicolon and 40.4% (n = 186) rectum; 14.6% (n = 67) had been with household historical past of CRC. There was no correlation between incidence age and intercourse (P = 0.06), tumor stage (P = 0.10), tumor location (P = 0.13), and household historical past of CRC (P = 0.3). We noticed a weak correlation between physique mass index (BMI) and age (Pearson correlation coefficient = 0.12, P = 0.01).
a The variety of sufferers within the Guangzhou cohort stratified by age and intercourse. b Two-dimension scatter plot exhibits the general sample of samples. Precept coordinate evaluation (PCoA) was carried out based mostly on the Bray–Curtis distance calculated from the abundance profile at species degree. Every level represents one pattern, and shade scale signifies age. Samples from feminine and male sufferers are in triangles and squares, respectively. Scatterplots of relationship between age and PCoA axis 1 (c), PCoA axis 2 (d), variety of species (e), and Shannon index (f). The correlation coefficient was calculated utilizing the Spearman technique. The stable pink line was fitted by clean operate in R, and the grey space is the 95% confidence interval. The Shannon index was calculated based mostly on the abundance profile on the species degree.
Restricted affiliation between the intestine microbiome and age in CRC sufferers
The connection between age and the intestine microbiome in CRC sufferers was investigated utilizing shotgun metagenomic sequencing of stool samples. We generated a complete of 32,403 million paired-end high-quality reads, with a median of 70 million paired-end reads per pattern (Strategies). We discovered no correlation between age and alpha-diversity, outlined because the variety of noticed species and Shannon index (Fig. 1e, f). The adjustment of confounders (BMI, intercourse, tumor location and stage, and smoking) didn’t improve the affiliation between alpha-diversity and age. Equally, there was no correlation between age and the primary two coordinates of the principal coordinate evaluation (PCoA) (Fig. 1b–d). This was supported by the permutation multivariate evaluation of variance (PERMANOVA) take a look at, displaying that age solely defined a small fraction of microbiome variance (R2 = 0.003, P = 0.15).
To determine particular taxa related to age, we examined the correlation between age and species abundance (Strategies). We solely discovered that the abundance of 4 species, specifically Prevotella stercorea, Bifidobacterium dentium, Prevotella copri, and Prevotella bivia, had been considerably correlated with age (false discovery fee (FDR) adjusted P < 0.05, Supplementary Knowledge 2). The associations of Prevotella species had been unfavorable, whereas that of B. dentium was constructive. B. dentium and P. bivia’s associations with age had been impartial of physique mass index (BMI), intercourse, tumor location and stage, household historical past of CRC, and smoking. Though Prevotella species are generally discovered within the human intestine microbiota and have been linked to dietary habits, their relative abundances had been age-dependent and dropped from maturity to outdated age20. B. dentium, which is usually discovered within the human oral microbiome21, had elevated abundance and prevalence within the intestine with age22. Whereas two dozen of different bacterial species have been recognized as age-associated23, their associations with age in CRC sufferers had been weak, suggesting that CRC standing could outperform age in shaping the intestine microbiome.
As CRC is likely one of the most studied traits in intestine microbiome analysis, a group of CRC-associated taxa has been robustly recognized in earlier research. We obtained an inventory of 118 CRC-associated taxa from gutMDisorder24 (Strategies). Amongst them, 24/25 taxa reported in at the very least two research had been detected in our cohort with prevalence charges from 15.43% to 90.65% (common 58.41%), with 16 taxa introduced in over half sufferers. Importantly, solely the abundance of P. copri was correlated with age, however its correlation was not important after adjustment for confounders (Supplementary Knowledge 2).
Moreover, we validated our findings in a not too long ago printed yCRC cohort (Fudan cohort)18. On this cohort, age was given as a binary variable, outdated or younger, with the cutoff of fifty. Constantly, we separated our Guangzhou sufferers into outdated (age ≥ 50) and younger (age < 50) teams. We solely discovered 9 species with differential abundance (P < 0.05) between young and old teams in each cohorts (Supplementary Knowledge 3). 4 of them (P. stercorea, B.dentium, P. copri and P. bivia) had been talked about above. Though the opposite 5 species included beforehand reported CRC-associated microbe Alistipes indistinctus10,15, and CRC-depleted microbes Eubacterium rectale10 and Faecalibacterium prausnitzii10, none handed the a number of testing correction (FDR adjusted P > 0.05). Taken collectively, there was restricted (if any) affiliation between the recognized CRC-associated taxa and age.
Bacterial species related to oCRC and yCRC
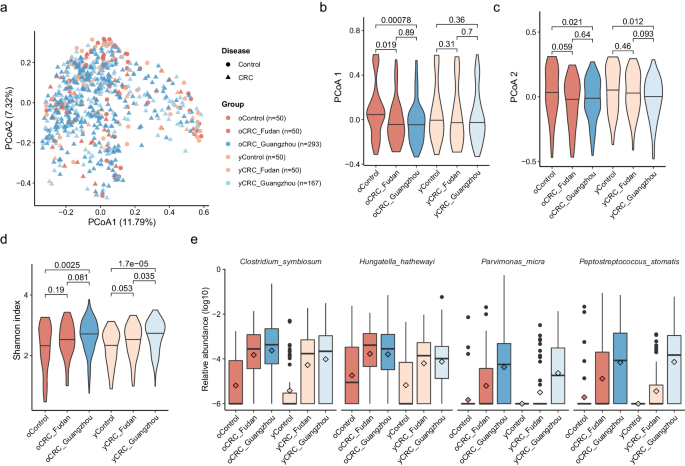
To research the intestine microbiome modifications in oCRC and yCRC sufferers, we in contrast them to age-matched controls. We reanalyzed the stool metagenomic information from the Fudan cohort18 and built-in it with our Guangzhou cohort (Strategies). In accordance with Yang et al.18, the yCRC was outlined as age beneath 50 years outdated, and the others had been oCRC. A PCoA based mostly on species-level abundance confirmed that illness impact surpassed the batch impact (Fig. 2a–c). The Guangzhou sufferers had related distributions in PCoA1 and PCoA2 with sufferers within the Fudan cohort, and decrease median values than the controls. Moreover, the CRC standing defined a barely larger variance than research impact (R2 = 0.00408 and 0.00376, PERMANOVA). Subsequently, the batch impact in Guangzhou and Fudan cohorts was restricted.
a Two-dimension scatter plot exhibits the general distribution of Fudan and Guangzhou samples. Precept coordinate evaluation (PCoA) was carried out based mostly on the Bray–Curtis distance calculated from the abundance profile at species degree. Every level represents one pattern. Samples from Fudan and Guangzhou cohorts are in pink and blue, respectively. Circles are management samples, whereas triangles are CRC samples. Violin plots present values of PCoA axis 1 (b) PCoA axis 2 (c), and Shannon index (d) throughout completely different teams. The thick horizon line signifies the 50% percentile. P values on the highest had been calculated by two-side Wilcoxon rank-sum take a look at. e 4 well-known CRC-enriched species considerably enriched in each oCRC and yCRC sufferers at false discovery fee (FDR) adjusted P < 0.05. The pattern measurement of every group is identical as a. The relative abundance is in log10 scale and zeros had been changed by a small worth. The field plots present the median (thick line), interquartile vary (field limits), 1.5× the interquartile vary span (whiskers), and outliers (dots). Diamond form signifies the imply abundance.
Earlier research have instructed that the microbiome of CRC sufferers had a better alpha range than controls, presumably because of the growth of usually oral microbes along with the baseline intestine microbiome13,14,17. We confirmed this discovering within the Fudan cohort, the place oCRC and yCRC sufferers had a better Shannon index than their controls (Fig. second). Guangzhou sufferers additionally had a better Shannon index than the Fudan controls, supporting the elevated range in CRC sufferers.
To determine taxa which can be differentially ample in CRC sufferers, we carried out two units testing. The primary set of testing was carried out solely on the Fudan cohort, whereas the second set was carried out on the Guangzhou sufferers and Fudan controls (Strategies). Our evaluation revealed 4 species (Clostridium symbiosum, Peptostreptococcus stomatis, Parvimonas micra, and Hungatella hathewayi) that had been constantly enriched (FDR adjusted P < 0.05) in oCRC and yCRC sufferers in each cohorts in comparison with Fudan controls (Fig. 2e, Supplementary Knowledge 4). These 4 species are well-known CRC-associated biomarkers13,14 and weren’t related to age (Supplementary Knowledge 3). C. symbiosum, as an example, was first reported by a qPCR research25 and confirmed in a meta-analysis research that built-in 5 shotgun metagenomic research13. P. micra and P. stomatis had been among the many most essential options in CRC classifiers constructed on stool microbiome information13.
We additionally discovered six different microbial species that had been differentially ample (FDR adjusted P < 0.05) in oCRC and confirmed related tendencies in yCRC in each cohorts (Supplementary Fig. 1, Supplementary Knowledge 4). Amongst these six taxa, Eggerthella lenta, Erysipelatoclostridium ramosum, and Flavonifractor plautii had been enriched in CRC teams and have been beforehand reported as biomarkers for CRC10,18. Particularly, F. plautii was recognized as a biomarker for yCRC by Yang et al.18. Two recognized micro organism Eubacterium rectale and Ruminococcus bicirculans, in addition to a metagenomically assembled taxon Eubacterium sp. CAG38 was depleted in CRC microbiome. E. rectale is likely one of the most prevalent human intestine micro organism26 and was reproducibly reported with decreased abundance in CRC sufferers in comparison with wholesome controls9,10.
Then again, three of the 4 taxa (Alistipes indistinctus, Clostridium aldenense, Eisenbergiella tayi, and Fusobacterium sp. oral taxon 370) enriched in yCRC confirmed related tendencies in oCRC as properly (Supplementary Fig. 2, Supplementary Knowledge 4). Fusobacterium sp. oral taxon 370 is likely one of the typical oral micro organism linked to CRC in old-onset sufferers14. Whereas A. indistinctus was at a low abundance within the human intestine microbiome, it was concerned in CRC carcinogenesis and remedy response27. We particularly analyzed two taxa, B. fragilis6 and F. nucleatum7, with proposed carcinogenesis mechanism. Though the B. fragilis abundance was larger in CRC than management, the importance was misplaced after a number of speculation adjustment (Supplementary Fig. 3). F. nucleatum’s prevalence was surprisingly low within the Fudan cohort, whereas its abundance had an identical distribution in oCRC and yCRC of the Guangzhou cohort. Moreover, on the important degree of nominal P < 0.05, 23 of 24 species passing the edge had directionally constant modifications in old- and young-onset sufferers in contrast with their controls (Supplementary Knowledge 4). Total, our findings point out that many of the CRC-associated taxa confirmed concordant modifications in each oCRC and yCRC microbiomes in comparison with their controls.
Pressure-level range of F. nucleatum, B. fragilis, and E. coli
Our deep metagenomics sequencing information allowed us to research the strain-level insights into CRC-associated species. Given the inherent challenges of pressure degree, we centered on three CRC-associated species F. nucleatum (Fn), B. fragilis (Bf), and E. coli (Ec) within the Guangzhou cohort. We used StrainPhlAn317 to assemble the phylogenetic tree and used inStrain28 to look at the genome-wide sequence range (Strategies).
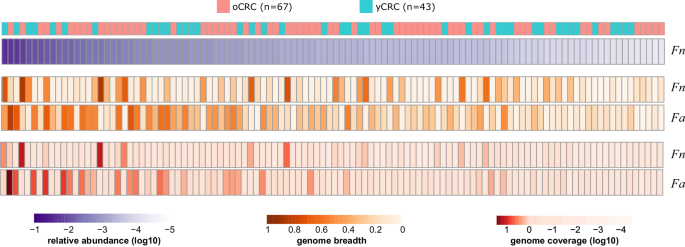
Fn was recognized in 63 samples based on its marker genes and the corresponding phylogenetic tree confirmed no correlation with affected person age, tumor stage, and placement (Supplementary Fig. 4a). No important distinction was noticed within the Fn prevalence between oCRC and yCRC. Genome-wide sequence evaluation revealed equally excessive population-level common nucleotide id (popANI)28 values to the Fn reference genome in oCRC and yCRC metagenomes, with no important distinction (Supplementary Fig. 4b). We additionally evaluated the nucleotide range, an indicator of pressure diversification. Our evaluation revealed no affiliation between Fn nucleotide range and affected person age, tumor stage or location (Supplementary Fig. 4c–e). A research reported that F. animalis (Fa, often known as Fn subspecies animalis) had larger abundance and prevalence than Fn in tumor samples29. In keeping with this discovering, we discovered Fa in additional samples with larger protection than Fn in our cohort (Fig. 3). Fa prevalence, popANI, and nucleotide range values confirmed no distinction in oCRC and yCRC (Supplementary Fig. 5). Taken collectively, the evaluation of Fn and Fa distribution and variety revealed no distinctions between oCRC and yCRC. Nonetheless, we famous a better nucleotide range of Fa within the colon in comparison with rectum tumors (Supplementary Fig. 5). An analogous pattern was noticed for Fn, albeit with decrease significance because of a smaller pattern measurement. This statement means that Fn and Fa exhibited elevated range in sufferers with colon tumors.
Heatmap exhibits the abundance, genome-wide breadth, and protection of F. nucleatum (Fn) and F. animalis (Fa). Solely 110 samples which had genome breadth >0.1 and protection >0.1 for Fn (RefSeq GCF_008633215.1) or Fa (RefSeq GCF_000158275.2) reference genomes had been included. Samples had been sorted in lowering order by the relative abundance of Fn, calculated by MetaPhlAn 3.
For Bf, we recognized two distinct phylogenetic clusters (Supplementary Fig. 6a). Cluster 1 (N = 267) was dominated by samples with metagenome-assembled genomes (MAGs) annotated to pressure NCTC9343 (common nucleotide id (ANI) > 95%), whereas cluster 2 (N = 67) was dominated by samples with MAGs annotated to pressure Q1F2 (Strategies). The distribution of oCRC and yCRC was not completely different within the two clusters. The phylogenetic tree revealed no correlation with affected person age, tumor stage or location, inside or between clusters (Supplementary Fig. 6a). Genome-level evaluation demonstrated excessive popANI values to the reference genomes in oCRC and yCRC (Supplementary Fig. 6b). Whereas the nucleotide range of cluster 1 was not related to age, the nucleotide range of cluster 2 was larger in yCRC than oCRC (P = 0.02, Supplementary Fig. 6c). This implies that Bf pressure in cluster 2 samples could also be beneath stronger choice stress in yCRC sufferers.
For E. coli, our evaluation recognized just one pressure cluster (ANI > 95% to pressure ATCC 11775) in 317 (69%) samples, with no important distinction in prevalence between oCRC and yCRC. The marker gene-based phylogenetic tree evaluation revealed no correlation with affected person age, tumor stage or location (Supplementary Fig. 7a). Genome-level evaluation indicated an identical popANI worth to the reference genome in oCRC and yCRC (Supplementary Fig. 7b). Nucleotide range evaluation confirmed no affiliation with affected person age, tumor stage or location (Supplementary Fig. 7c–e).
Useful metagenomic signatures for oCRC and yCRC
Not like 16S rRNA gene amplicon information, metagenomes permit us to entry the useful capability of the intestine microbiome. In our Guangzhou cohort, age didn’t have a big affiliation with the highest two axis of PCoA, calculated from the microbial pathway profile (Supplementary Fig. 8). In PERMANOVA, age defined a small and non-significant quantity of general variance of the microbial pathway variation (R2 = 0.005, P = 0.13). To determine particular microbial pathway related to age, we examined the associations between age and abundances of metaCyc30 pathways (Strategies). Just one metaCyc pathway, PWY-6608: guanosine nucleotides degradation III, was related to age at P < 0.01 with and with out adjustment for confounding elements (Supplementary Knowledge 5).
To determine useful signatures related to oCRC and yCRC, we built-in the useful pathway profiles of our Guangzhou cohort and the printed Fudan cohort. PCoA confirmed that Guangzhou sufferers had been nearer to Fudan sufferers than controls (Supplementary Fig. 9). As well as, CRC standing defined larger variance than the research impact (R2 = 0.014 vs. 0.006, PERMANOVA). We then carried out differential evaluation in metaCyc pathways (Strategies). Out of the 435 examined metaCyc pathways, 69 had differential abundance between oCRC and age-matched controls in each cohorts (FDR adjusted P < 0.05, Supplementary Knowledge 6). Amongst these, whereas just one pathway, PWY-7316: dTDP-N-acetylviosamine biosynthesis, was differential in yCRC and their controls on the identical important degree, the bulk (60/69) had concordant enrichment path in yCRC.
We subsequent examined the well-known CRC-associated microbial cutC gene, which encodes choline trimethylamine-lyase liable for manufacturing of the disease-associated trimethylamine (TMA). In whole, 113 UniRef9031 gene households annotated as cutC orthologs had been detected in at the very least one pattern in Guangzhou or Fudan cohorts. Of those, 8 cutC orthologs introduced in additional than half of Guangzhou sufferers had concordant enriched path in oCRC and yCRC in comparison with their controls (Supplementary Knowledge 7). Remarkably, two cutC orthologs with represented sequences from Bacteroides species, together with Bf ), reached a statistically important degree of nominal P < 0.05. The sum abundance of cutC gene households was larger in oCRC and yCRC in comparison with their controls (Supplementary Fig. 10).
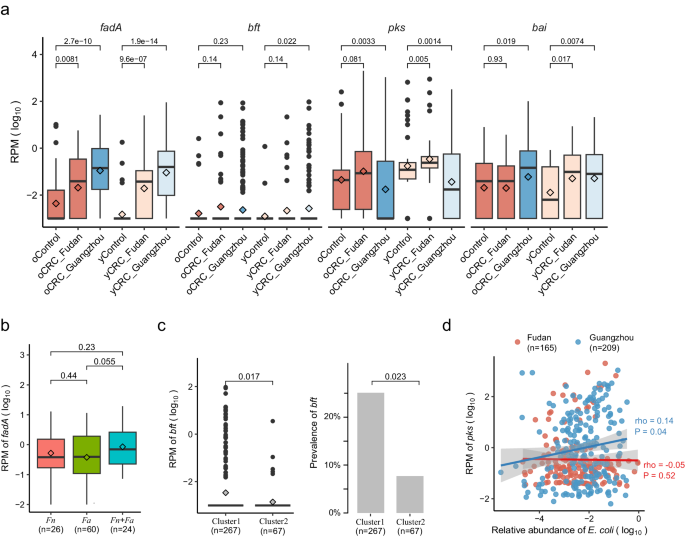
We accessed CRC-associated virulence elements and toxins, specializing in fadA (encodes Fn adhesion protein A)7, bft (encodes Bf enterotoxin)5, the pks genomic island (encodes colibactin in some Ec strains)8, and the bai operon (encodes enzymes for the conversion of main to secondary bile acids in Clostridium species)32 (Strategies). fadA exhibited important enrichment in each oCRC and yCRC in comparison with their respective controls (Fig. 4a). Within the strain-level evaluation, we recognized Fn and Fa in our samples. Right here we additional explored fadA abundance within the context of Fn and Fa. fadA abundance was not completely different in samples with both Fn or Fa (Fig. 4b). Samples with each Fn and Fa had the very best fadA common abundance, whereas samples with none pressure exhibited the bottom fadA common abundance. For bft, we noticed an enrichment pattern in CRC in comparison with controls (Fig. 4a). Notably, each bft abundance and prevalence had been differentiated within the two phylogenetic clusters outlined within the strain-level evaluation part (Fig. 4c). The pks abundance exhibited variability between cohorts, being larger in CRC than controls within the Fudan cohort however decrease within the Guangzhou CRC than controls (Fig. 4a). Pressure-level evaluation recognized pressure Ec NCTC9343 was the dominant pressure within the Guangzhou cohort, and the reference genome of this pressure didn’t comprise the pks island. We explored the correlation between Ec and pks abundances, discovering a constructive correlation within the Guangzhou cohort however no correlation within the Fudan cohort (Fig. 4d). This discrepancy suggests a possible distinction within the ecological context between populations. For bai, its abundance was considerably larger in yCRC than within the respective controls (Fig. 4a). Within the Fudan cohort, oCRC and their controls had related ranges of bai, which had been larger than these in younger controls. This discovering aligns with stories of elevated bile acid metabolism in aged folks33, indicating that getting older could affect the affiliation between bai and CRC within the aged. In abstract, CRC-associated virulence elements (fadA, bft) had been enriched in each oCRC and yCRC.
a Normalized log abundance of CRC-associated virulence elements in several teams. RPM means reads per million mapped reads, see Strategies for gene quantification. fadA encodes F. nucleatum adhesion protein A; bft encodes B. fragilis enterotoxin; the pks genomic island encodes enzymes to provide genotoxic colibactin (in E. coli); the bai operon encodes bile acid-converting enzymes (current in some Clostridiales species). Pattern sizes for the in contrast teams: oControl (n = 50), oCRC_Fudan (n = 50), oCRC_Guangzhou (n = 293), yControl (n = 50), yCRC_Fudan (n = 50), yCRC_Guangzhou (n = 167). b Normalized log abundances of fadA stratified by the presence of F. nucleatum (Fn) and F. animilis (Fa). Presence was outlined as genome breadth >0.1 and protection >0.1. c Normalized log abundance and prevalence of bft stratified by B. fragilis pressure clusters. Cluster task was carried out based mostly on marker genes and genome-wide sequence evaluation (Strategies). P values on the highest had been calculated by two-side Wilcoxon rank-sum take a look at. d Correlation between normalized log abundances of pks and E. coli. The correlation coefficient was calculated utilizing the Spearman technique. The stable (blue and pink) traces had been fitted by clean operate in R, and the grey space is the 95% confidence interval. The boxplot conventions are in line with the outline in Fig. 2.
Associations between microbial markers and CRC traits
CRC is heterogenous and has various molecular traits. Right here, we investigated the associations between 18 taxonomic markers (these highlighted in Fig. 2, Supplementary Figs. 1–3) and CRC molecular traits, together with tumor stage and location, mismatch restore (MMR), BRAF and HER2 mutation standing (Strategies, Supplementary Knowledge 8). Larger abundance of P. stomatis was noticed in stage III sufferers, whereas E. rectale confirmed elevated abundance in stage II sufferers, each in comparison with stage I sufferers. Moreover, the abundance of E. ramosum was decrease in sufferers with rectal tumors, E. rectale was larger in sufferers with left-side tumors, P. micra was larger in sufferers with rectal tumors, relative to their counterparts with right-sided tumors. Nonetheless, in our pattern evaluation, we didn’t observe any statistically important monotonic relationships of those 18 taxa regarding tumor stage and placement.
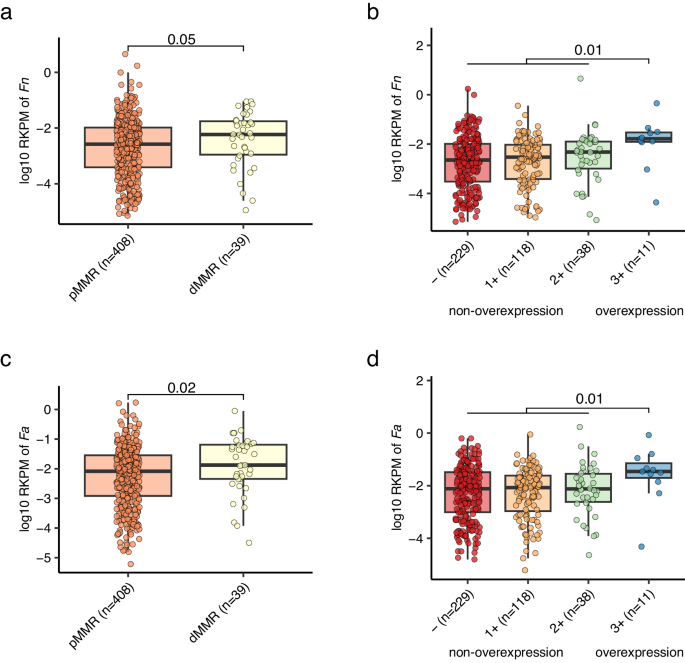
Notably, the relative abundance of Fn (maker-gene-based quantification) was larger in MMR poor (dMMR) sufferers than MMR proficient (pMMR) sufferers, and better in sufferers with HER2 overexpression in comparison with these with out overexpression (Supplementary Knowledge 8). We additional investigated such associations utilizing genome-based quantification, which may supply strain-level decision as described within the previous part. Intriguingly, each Fn and Fa demonstrated elevated abundance in sufferers with dMMR and HER2 overexpression, relative to their counterparts (Fig. 5).
The abundances of Fn (a) and Fa (c) had been larger in dMMR sufferers in comparison with pMMR sufferers. The abundances of Fn (b) and Fa (d) had been larger in HER2 overexpression sufferers in comparison with non-overexpression sufferers. Fn and Fa abundances had been decided by genome-wide reads mapping to their reference genome (Strategies). MMR and HER2 standing had been decided by immunohistochemistry take a look at. RKPM: reads per kilobase per million reads. P values on the highest had been calculated by two-side Wilcoxon rank-sum take a look at. The boxplot conventions are in line with the outline in Fig. 2.
We moreover examined the associations between gene markers (fadA, bft, pks, and bai) and tumor stage, location, in addition to MMR, BRAF, and HER2 mutation standing. No important affiliation was recognized in our cohort.
Comparable prediction accuracy of CRC standing in old- and young-onset sufferers
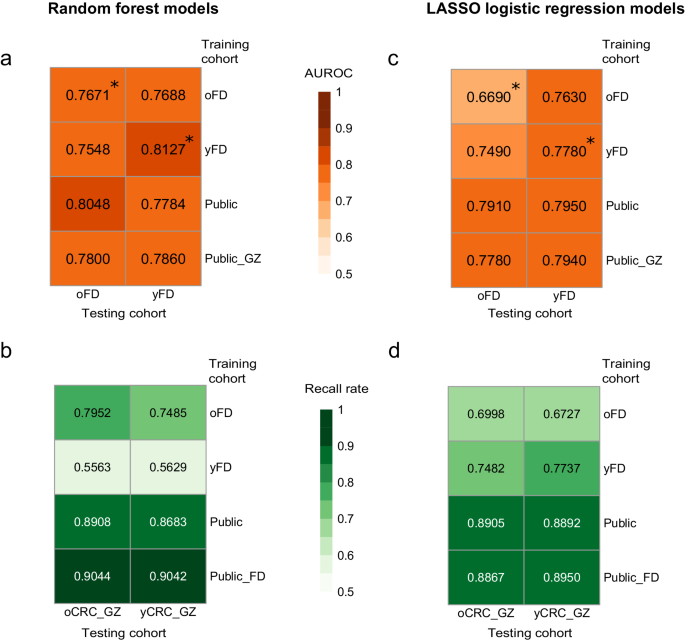
A number of research have demonstrated the potential of tailoring the intestine microbiome for predicting CRC standing13,14,17. To entry the transferability of classifiers throughout completely different affected person teams, we employed the random forest algorithm to coach machine studying fashions individually on oCRC and yCRC sufferers (Strategies). Our outcomes revealed promising cross-application efficiency. The mannequin skilled on Fudan oCRC sufferers exhibited sturdy predictive functionality for Fudan yCRC sufferers, attaining an space beneath receiver operator curve (AUROC) of 0.7688, solely barely decrease than the cross-validated AUROC of 0.8127 (Fig. 6a). Equally, the mannequin skilled on Fudan yCRC sufferers carried out equally to the cross-validation on oCRC sufferers, with an AUROC of 0.7548 and 0.7671, respectively (Fig. 6a). We prolonged our analysis to the Guangzhou cohort. The oCRC mannequin demonstrated a excessive recall fee of 0.7952 when predicting CRC standing in Guangzhou oCRC sufferers, and surprisingly, it outperformed the yCRC mannequin in predicting CRC standing in Guangzhou yCRC sufferers (0.7485 vs. 0.5629, Fig. 6b).
Prediction efficiency on oCRC and yCRC within the Fudan cohort and Guangzhou cohort utilizing fashions skilled on species-level abundances from completely different datasets. Fashions had been skilled on two completely different strategies: random forest and LASSO logistic regression. The numerical values are the realm beneath receiver operator curve (AUROC) for a and c, and the recall fee for b and d. Asterisks denote the values averaged over 100 occasions of 10-fold cross-validation. oFD means the 100 metagenomes of Fudan oCRC and oControl; yFD means the 100 metagenomes of Fudan yCRC and yControl; Public means 1262 public metagenomes; Public_GZ means 1262 public metagenomes plus 460 Guangzhou metagenomes; Public_FD means 1262 public metagenomes plus 200 Fudan metagenomes.
To beat the limitation of small pattern measurement within the Fudan cohort, we integrated the publicly out there dataset that consisted of 600 CRCs and 662 controls, which included solely 72 sufferers recognized beneath the age of fifty9,10,11,12,13,14,15,16. The mannequin skilled on this expanded dataset predicted CRC standing barely higher in Fudan oCRC sufferers in comparison with yCRC sufferers (AUROC = 0.8048 vs. 0.7784, Fig. 6a). Equally, the mannequin predicted CRC standing within the Guangzhou cohort with a barely larger recall fee in oCRC than yCRC sufferers (0.8908 vs. 0.8683, Fig. 6b). Once we included Guangzhou CRC sufferers within the coaching information, the ensuing mannequin had related prediction accuracy for oCRC and yCRC sufferers within the Fudan cohort (AUROC = 0.7800 and 0.7860, Fig. 6a). The mannequin skilled on the general public and Fudan datasets additionally confirmed related efficiency on oCRC and yCRC sufferers within the Guangzhou cohort (recall fee=0.9044 and 0.9042). In abstract, our outcomes counsel that the microbiome-based classifiers can predict CRC standing in each old- and young-onset sufferers with related accuracy.
Moreover, we skilled the random forest fashions utilizing metagenomic pathway profiles to foretell CRC standing (Strategies). The general efficiency of the pathway-based mannequin, as measured by the AUROC and recall fee, was decrease than that of the species-based mannequin (Supplementary Fig. 11). This aligns with earlier research reporting that metagenomic pathway-based CRC prediction fashions are likely to exhibit comparatively poorer efficiency in comparison with species-based fashions13,17.
Lastly, we replicated our machine studying experiments utilizing one other technique, least absolute shrinkage, and choice operator (LASSO) logistic regression. In line with the findings from the random forest strategy, the LASSO fashions constructed on species profiles carried out higher than the pathway profiles (Fig. 6c, d, Supplementary Fig. 11c, d). Importantly, the efficiency of LASSO fashions on oCRC and yCRC was related, suggesting the fashions’ transferability throughout completely different age teams.