Identification of subtypes and building of the LFMRS in AML
To discover whether or not lipid metabolism is said to the progress and prognosis of AML, we first carried out the survival analyses of 4 fatty acid metabolism-related gene units (KEGG, Hallmark, Reactom and WikiPathway) quantified by single pattern gene set enrichment evaluation (ssGSEA), and located that the prognosis of AML sufferers with excessive fatty acid metabolism is worse (Fig. 1A–D). Afterwards, all of the genes associated to fatty acid metabolism within the 4 fatty acid metabolism-related gene units have been collected as proven in Fig. S1A. As LSCs management the recurrence and refractory of AML sufferers, the fatty acid metabolism-related genes which extremely expressed in LSCs could also be a possible therapeutic goal. Thus, we urge to hunt the differentially expressed genes between LSCs and HSCs, and the three datasets of GSE68172 (5 HSC samples and 19 LSC samples), GSE17054 (4 HSC samples and 9 LSC samples), and GSE24395 (5 HSC samples and 12 LSC samples) have been utilized to dig up the potential effectors by strong rank aggregation technique. (Fig. S1B). As proven, a complete of 211 differentially expressed genes in LSCs have been enriched, together with 109 up-regulated genes and 102 down-regulated genes (Fig. 1E). Persistently, we carried out operate evaluation to analyze the organic processes and the corresponding pathways of differentially expressed genes in LSCs by Metascape, and located some lipid metabolism-related organic processes, comparable to “glycerophospholipid metabolic course of” in up-regulated differentially expressed genes (Fig. 1F) and “lipid homeostasis course of” in down-regulated differentially expressed genes (Fig. 1G). Thus, we screened the potential regulators of fatty acid metabolism in LSCs by intersecting the differentially expressed genes in LSCs (Fig. 1E) with the genes associated to fatty acid metabolism (Fig. S1A), and 9 potential genes have been based (Fig. 1H). The prognostic significance of 9 potential genes in TCGA-LAML have been analyzed by univariate COX regression, and 6 genes (LGALS1, ALDH1A1, AADAT, ELOVL7, ACOX2, and ACSM3) have been considerably correlated with the prognosis of AML sufferers. LGALS1, ALDH1A1, AADAT, ELOVL7, and ACOX2 have been the danger elements for the prognosis of AML sufferers, whereas ACSM3 was the protecting issue (Fig. 1I).
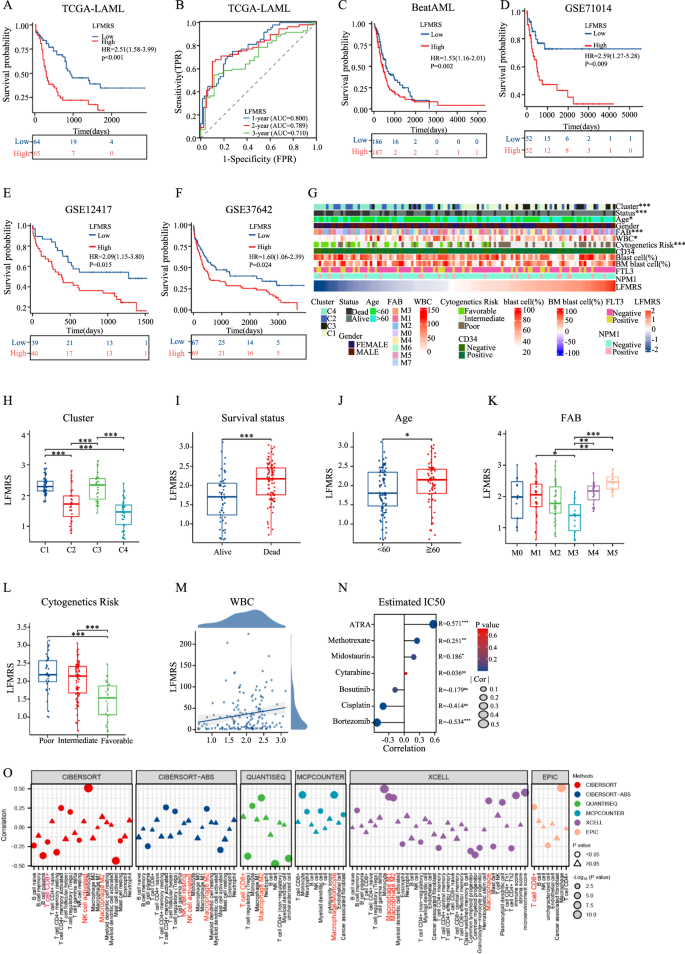
The survival analyses of ssGSEA rating of fatty acid metabolism-related gene units from KEGG (A), Hallmark (B), Reactom (C) and Wp (D) within the TCGA-LAML cohort. E The differential expression genes between LSCs and HSCs within the three databases (GSE17054, GSE68172, GSE24395) have been proven in a heatmap. Purple represents the considerably upregulated genes in LSCs in contrast with HSCs. Blue represents the considerably downregulated genes in LSCs in contrast with HSCs. Operate and pathway enrichment evaluation of the considerably upregulated genes (F) and downregulated genes (G) in LSCs versus HSCs by Metascape. The picture reveals the histogram of the highest 20 enriched pathway. H The Venn diagrams have been used to display the differential expression associated with fatty acid metabolism in LSCs. I Univariate COX regression evaluation of the 9 potential genes in TCGA-LAML. J Consensus matrix when ok = 4. Okay Kaplan-Meier OS curves for AML sufferers amongst C1, C2, C3, and C4 in TCGA-LAML. This desk beneath Kaplan-Meier OS curves reveals that the remaining sufferers who do not need the top level occasion (dying) beneath the indicated time on this subgroup. They’re susceptible to an endpoint occasion, which is known as quantity in danger. L The expression of LFMGs amongst C1, C2, C3, and C4. M Heatmap of correlation between lipid metabolism-associated genes in LSCs with clinicopathological traits of AML sufferers within the TCGA-LAML cohort. N Lasso COX regression evaluation of 5 OS-related genes. O The general survival (OS) within the TCGA-LAML cohort database was analyzed by the univariate COX regression with the 5 potential genes and summarized in Forest plots. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001; ns, not vital.
Subsequent, we carried out the consensus clustering evaluation utilizing R bundle “ConsensusClusterPlus” with cluster variable vary sited from 2 to 10 based mostly on these six potential effectors to establish the potential fatty acid metabolism-related subtypes in AML sufferers. First, we carried out cumulative distribution operate (CDF) plots, and located that when ok = 4, the descending slope of CDF is smaller in contrast with ok = 2, ok = 3, and ok = 5, and CDF reached an approximate most (Fig. S1C). Additionally, delta space plot was made. As proven in Fig. S1D, when ok = 5, the change of space beneath curve was slighter than ok = 4. In short, 4 clusters are appropriate for the best intragroup correlations and the bottom intergroup correlations, which might be noticed in Fig. S1E. These information indicated that ok = 4 confirmed distinguished clustering stability with the best intragroup correlations and the bottom intergroup correlations. Thus, we acquired 4 clusters (C1, C2, C3, and C4) (Fig. 1J). As well as, the clusters have been validated by PCA evaluation (Fig. S1F). Moreover, we analyzed the survival of C1, C2, C3, and C4 clusters, and located that C1 had the worst prognosis and C4 had one of the best prognosis (C1 < C3 < C2 < C4) (Fig. 1K). Collectively, the relative expression of the six potential effectors have been detected, and located most danger elements (LGALS1, ALDH1A1, ELOVL7 and ACOX2) have been excessive expressed in C1 and C3 clusters, and the protecting issue (ACSM3) have been excessive expressed in C2 and C4 clusters (Fig. 1L). Subsequent, we investigated the affiliation of LSCs and fatty acid metabolism-related clusters with classical medical options of AML within the TCGA-LAML cohort, LSCs and fatty acid metabolism related-clusters, survival standing, general survival, age, intercourse, FAB typing, WAB quantity, cytogenetic danger, CD34, proportion of progenitor cells, proportion of precursor cells in bone marrow, FLT3 gene mutation, NPM1 mutation, and expression profile of every gene have been used as annotations (Fig. 1M). The outcomes confirmed that the survival standing was poorer (Fig. S1G), the share in M5 (the worst prognosis subtype) [24] (Fig. S1H), WBC counts (Fig. S1I), and poor cytogenetics danger (Fig. S1J) have been greater in C1 and C3 clusters (worst prognosis) in contrast with C2 and C4 clusters, which indicated that the 4 clusters have been efficiently recognized based mostly on the potential six genes of AML.
To raised help clinicians in precisely predicting the prognosis of AML sufferers, we attempt to assemble the LSCs and fatty acid metabolism-related danger rating (LFMRS) based mostly on the six potential genes. The Least Absolute Shrinkage and Choice Operator (LASSO) regression algorithm decided 5 OS-related genes based mostly on the optimum λ worth and the minimal partial chance of deviance (Fig. 1N, S1K). LASSO coefficients of the 5 potential genes confirmed that LGALS1 (0.260), ELOVL7 (0.215), ALDH1A1 (0.045), and ACOX2 (0.013) have been danger elements and ACSM3 (-0.138) was a protecting issue (Fig. 1O).
Verification of prognostic mannequin (LFMRS) in AML
In response to the median danger rating of LFMRS in coaching cohort of TCGA, AML sufferers have been divided into low-risk group and high-risk group (Fig. S2A). Not surprisingly, sufferers in high-risk group had shorter survival time and worse prognosis than these in low-risk group (Fig. 2A). Furthermore, the sensitivity and specificity of LFMRS have been estimated by time-dependent receiver working attribute (ROC) evaluation, and the areas beneath the curve (AUCs) for one-year, two-year, and three-year general survival have been 0.800, 0.789, and 0.710, respectively with vital p values (Fig. 2B). For verifying the reliability of LFMRS, the identical evaluation is carried out within the 4 databases of BeatAML (Fig. 2C, S2B), GSE71014 (Fig. 2D, S2C), GSE12417 (Fig. 2E, S2D), and GSE37642 (Fig. 2F, S2E), and the outcomes present that the constructed LFMRS has a excessive prognostic reliability.
A Kaplan-Meier evaluation of OS between high- and low-LFMRS teams within the TCGA cohort. B The 1-, 2- and 3-year ROC curves of the LFMRS within the TCGA cohort. Kaplan-Meier analyses of OS between high- and low-LFMRS teams within the BeatAML database (C), GSE71014 (D), GSE12417 (E) and GSE37642 (F) cohorts. G Overview of the correspondence between LFMRS and different medical options of AML sufferers. LFMRS expression amongst distinct clusters (H), between alive and useless sufferers (I), between <60 and ≥60 sufferers (J), amongst distinct FAB subtypes (Okay), amongst totally different cytogenetic dangers (L) of AML. M The correlation of LFMRS with WBC counts. N The correlation of LFMRS with IC50 values of first-line medication from GDSC database quantified by pRRophetic. O Correlation of LFMRS with immune cells quantified by CIBERSORT, CIBERSORT-ABS, QUANTISEQ, MCPCOUNTER, XCELL and EPIC in AML. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001; ns not vital.
Subsequent, we stratified the AML sufferers into high-risk and low-risk teams in line with their LFMRS scores and assessed their medical parameters, and located that the distribution of the clusters, survival standing, age, FAB typing, WBC counts, and cytogenetic risk-based danger teams have been totally different between the LFMRS high- and low-risk teams, whereas different medical options confirmed no significance (Fig. 2G). We additionally analyzed the LFMRS danger values among the many clusters, survival standing, age, FAB classification, WBC counts, and cytogenetic danger, and located that LFMRS have been greater in sufferers with C1 and C3 clusters (Fig. 2H), poor survival standing (Fig. 2I), older than 60 years outdated (Fig. 2J), M5 sort in FAB classification (Fig. 2K), and poor cytogenetic danger (Fig. 2L). LFMRS danger values was positively correlated with WBC counts (Fig. 2M). Furthermore, we detected the correlation between LFMRS and estimated drug IC50 quantified by pRRophetic technique (Fig. S2F), and located that LFMRS was probably the most delicate to parthenolide and least delicate to ATRA in AML (Fig. S2G). The identical end result was additionally discovered by the evaluation of the correlation between medical utilized medication in AML and LFMRS, which indicating that LFMRS often is the key issue of drug resistance, particularly in retinoic acid remedy of AML (Fig. 2N). In the meantime, we additionally assessed the correlation between LFMRS and immune cells utilizing CIBERSORT, CIBERSORT-ABS, QUANTISEQ, MCPCOUNTER, XCELL, and EPIC algorithms, and located that macrophage, CD8+ T cells, NK cell, et. al. have been carefully associated to LFMRS (Fig. 2O). Thus, these information indicated that LFMRS danger classification have been in line with present danger elements.
To find out whether or not the LFMRS is independently correlated with the OS of AML sufferers, we first carried out univariate COX regression evaluation. By means of analyzing the prognostic worth of LFMRS along with different widespread prognostic elements (age, gender, WBC counter, cytogenetics danger, CD34 expression, the share of blast cell and BM blast cell, FLT3 mutation, and NPM1 mutation), we discovered that age, cytogenetic danger and LFMRS have been related to prognosis of sufferers with AML (Fig. S3A). Moreover, we additionally carried out multivariate COX regression evaluation, and the outcomes additionally confirmed that age and LFMRS have been unbiased prognostic elements (Fig. S3B). Subsequent, we established a prognostic nomogram integrating age and LFMRS (Fig. S3C), and the calibration curve of nomogram confirmed excessive concordance between the anticipated and precise chances of 1-, 2- and 3-year survival (Fig. S3D). The reliability of the predictive impact of the nomogram on prognosis have been additional verified in 1-, 2-, and 3-year prognosis by ROC curves (Fig. S3E). Total, these outcomes indicated that LFMRS is an unbiased danger issue, might be mixed with age for higher exactly predicting the survival time of AML sufferers.
LGALS1 is very expressed in LSCs and related to poor prognosis of AML sufferers
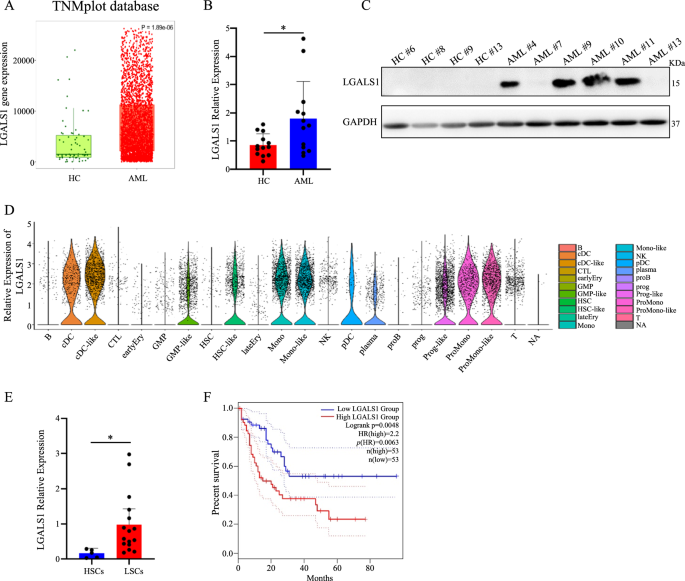
LGALS1 is the one member of LFMRS mannequin which is very expressed in LSCs in contrast with HSCs (Fig. 1E), has probably the most vital prognostic impact with the best HR and the bottom p worth (Fig. 1I). and is the molecule with the best prognosis danger (Fig. 1O). Moreover, LGALS1 is very expressed in worst-prognosis cluster C1 in AML (Fig. 1L) and is positively correlated with LFMRS rating (Fig. S4A). Thus, LGALS1 caught our consideration to additional discover the operate and mechanism AML to confirm LFMRS mannequin. To validated whether or not LGALS1 is very expressed in LSCs, we first analyzed LGALS1 expression by TNMplot database (Fig. 3A) and TARGET database (Fig. S4B), and located that LGALS1 was excessive expressed in AML sufferers than wholesome people. In the meantime, we collected bone marrow samples from AML sufferers, additionally discovered that LGALS1 was aberrantly overexpressed in AML samples relative to wholesome controls at each the mRNA degree (Fig. 3B) and protein degree (Fig. 3C). Subsequent, we analyzed the one cell sequencing information from GSE116256, and located that LGALS1 expression was greater in LSC-like (HSC-like and GMP-like) in contrast with HSC-like (HSC and GMP) (Fig. 3D). Additionally, we sorted LSCs from AML sufferers and HSCs in particular person of wholesome, and located that LGALS1 expression was greater in LSCs than HSCs (Fig. 3E). Furthermore, the elevated expression of LGALS1 correlated with poor survival of AML sufferers (Fig. 3F). Thus, LGALS1 is very expressed in LSCs and related to poor prognosis of AML sufferers.
A The transcript ranges of LGALS1 in AML samples in contrast with that in wholesome people have been recognized from TNMplot database. B The mRNA ranges of LGALS1 in major AML circumstances (n = 13, AML#1-AML#13) and wholesome management circumstances (n = 13). C The protein ranges of LGALS1 in indicated major AML circumstances and wholesome management circumstances. D The relative mRNA expression of LAGLS1 from the one cell sequencing information of GSE116256. E The mRNA ranges of LGALS1 in HSCs and LSCs from wholesome management circumstances and AML circumstances, respectively (LSCs from AML#1-AML#16). F Kaplan-Meier plots of general survival in TCGA cohorts for AML sufferers, stratified on the idea of LGALS1 expression above (LGALS1excessive) or under (LGALS1low) the median. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001; ns, not vital.
As well as, we additionally assessed the expression and prognosis of others members of LFMRS mannequin. As proven in Fig. S4C, ACSM3 was excessive expressed in AML than well being particular person, whereas ELOVL7 and ALDH1A1, and ACOX2 have been low expressed by TARGET database. Furthermore, there have been no vital distinction of ACSM3 and ACOX2 expressions between AML samples relative to wholesome controls, whereas ELOVL7 and ALDH1A1 have been aberrantly low-expressed in our gathering samples (Fig. S4D). We additionally revealed that ELOVL7 and ACOX2 have been decrease in LSCs in contrast with HSCs, and there have been no vital distinction of ALDH1A1 and ACSM3 expressions between two teams (Fig. S4E). The excessive expression of ELOVL7, ALDH1A1, and ACOX2 correlated with poor survival of AML sufferers, whereas excessive expression of ACSM3 associated with good survival of AML sufferers (Fig. S4F). These outcomes are principally in line with LFMRS mannequin.
LGALS1 promotes cell proliferation and inhibits cell apoptosis
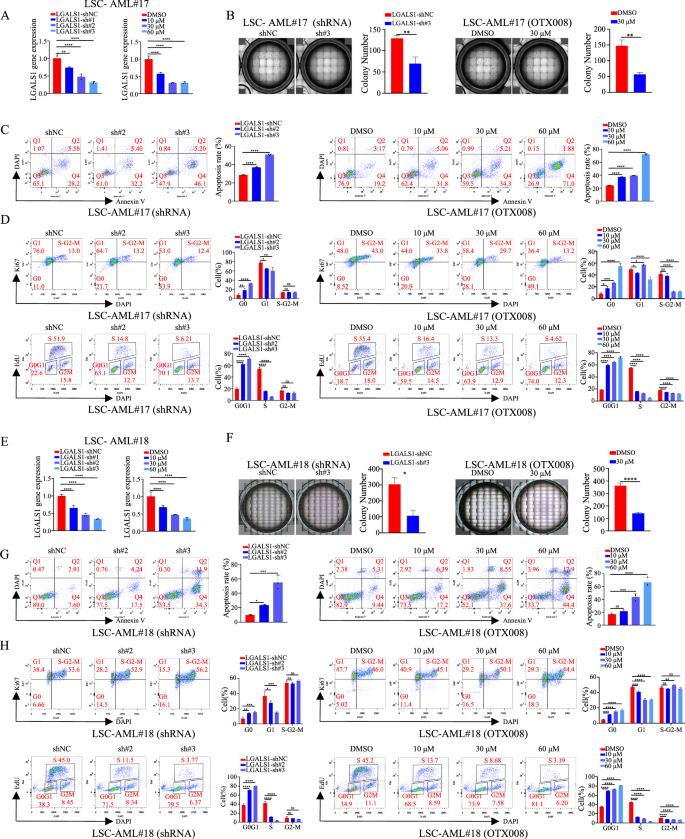
To discover the operate of LGALS1 in LSCs, we sorted LSCs (CD34+CD38−) from two AML sufferers. As proven in Fig. 4A–H, depletion of LGALS1 expression by shRNA (Figs. 4A, E) or inhibiting LGALS1 expression (Figs. 4A, E) utilizing OTX008 (a particular inhibitor of LGALS1) [25] impaired cell proliferation through colony formation assay (Figs. 4B, F), enhanced cell apoptosis through stream cytometric evaluation (Figs. 4C, G), and led to a lower within the fraction of cells in S section (offered the speed of proliferation) and improve in that in G0 section (Figs. 4D, H). We additionally decided LGALS1 expression in a set of leukemia cells, and located a relative stronger endogenous LGALS1 was seen in HEL, THP1, MV411, and NB4 cells (Fig. S5A, S5B). Subsequently, we explored the operate in AML utilizing HEL and THP1 cells, and located that suppression of LGALS1 (Fig. S5C, S6A) in THP1 and HEL cells additionally impaired cell proliferation (Fig. S5D, S6B), enhanced cell apoptosis (Fig. S5E, S6C), and led to a lower within the fraction of cells in S section (Fig. 5F, S6D). Thus, LGALS1 promotes cell proliferation and inhibits cell apoptosis in LSCs and leukemia cells in vitro.
A–D LSCs have been sorted from AML#17. A Efficiencies of LGALS1 silence in LSCs have been decided by qRT-PCR. B Cell development was decided by colony formation assay beneath a lightweight microscope, and the share of colony formation models have been proven. C Cell apoptosis was decided by stream cytometric evaluation. D Cell cycle distribution was detected by stream cytometric evaluation through Ki67 staining (higher) and EdU staining (decrease), respectively, and the bar graph confirmed the share of G0/G1, S, and G2/M section cells. E–H LSCs have been sorted from AML#18. E Efficiencies of LGALS1 silence in LSCs have been decided by qRT-PCR. F Cell development was decided by colony formation assay beneath a lightweight microscope, and the share of colony formation models have been proven. G Cell apoptosis was decided by stream cytometric evaluation. H Cell cycle distribution was detected by stream cytometric evaluation through Ki67 staining (higher) and EdU staining (decrease), respectively, and the bar graph confirmed the share of G0/G1, S, and G2/M section cells. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001; ns not vital.
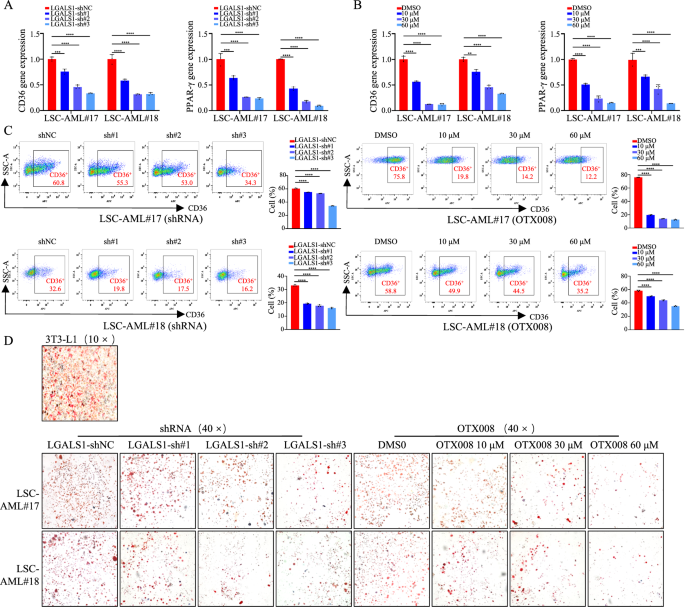
A–D LSCs transfected with shRNA in opposition to GALS1, or handled with DMSO or OTX008 have been cultured. A, B The mRNA ranges of CD36 and PPAR-γ have been detected by qRT-PCR. C The protein ranges of CD36 have been decided by FCM utilizing anti-CD36-APC (1:100, BioLegend, America). D Consultant photos of Oil Purple O staining. Differentiated 3T3-L1 cell was used as a optimistic management. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001; ns not vital.
LGALS1 contributes to lipid metabolism reprogramming
Lipid metabolism is regulated by a mix of the uptake and export of fatty acids, de novo lipogenesis, and fats utilization by β-oxidation [26]. It’s reported that LGALS1 is related to lipid synthesis in adipocyte by activation of peroxisome proliferator-activated receptor gamma (PPARγ) in adipose cell [26]. Galectin-1 has additionally been proposed to manage adipogenesis and adipose irritation by binding to CD146 [27]. Whereas, whether or not LGALS1 regulates lipid metabolism in leukemia cells stays unknown. Subsequently, we refine the impact of LGALS1 on the lipid uptake and de novo lipogenesis in LSCs and leukemia cells. First, we detected the expression of gene-related to lipid uptake (CD36) and de novo lipogenesis (PPAR-γ, FASN, ACC), and located that depletion of LGALS1 expression by shRNA or inhibiting LGALS1 expression utilizing OTX008 decreased CD36 and PPAR-γ expression (Fig. 5A–C, S7A–S7C), which indicated that LGALS1 might improve lipid accumulation in LSCs, HEL, and THP1 cells. Not surprisingly, Oil Purple O staining confirmed that restrained LGALS1 considerably diminished the buildup of lipid droplets in LSCs and leukemia cells (Fig. 5D, S7D). The above information point out that LGALS1 enhances the buildup of fatty acids in LSCs and leukemia cells.
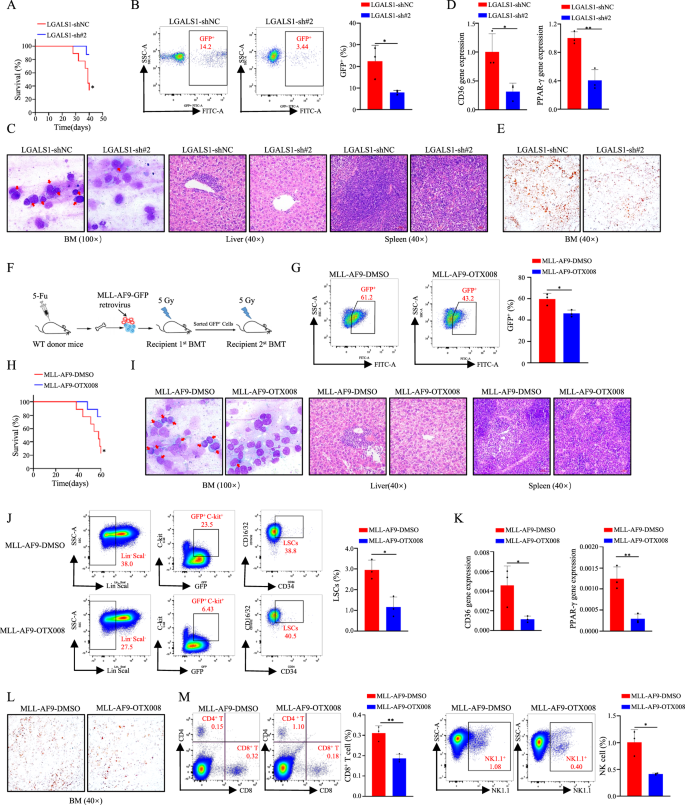
LGALS1 enhances lipid metabolism reprogramming, an immunosuppressive microenvironment, and AML development in vivo
To look at the mitogenic impact of LGALS1 in vivo, engineered HEL cells have been injected into NOD/SCID mice, as anticipated, we discovered that the mice injected with shLGALS1 cells confirmed longer survival time than these injected with shNC cells (Fig. 6A). LGALS1 knockdown inhibited leukemogenesis (Fig. 6B). Subsequent, leukemia cell infiltration was assessed. As proven in Fig. 6C, LGALS1 knockdown diminished leukemic cells within the bone marrow, and curbed liver and splenic infiltration as detected by H&E staining (Fig. 6C). Not surprisingly, the degrees of CD36, PPAR-γ, and lipid droplet have been diminished in xenografts from mice receiving LGALS1 knockdown cells (Fig. 6D, E). These outcomes recommend that LGALS1 regulates enlargement and lipid metabolism reprogramming of leukemic cells.
A Kaplan-Meier evaluation of the survival curves of the mice in every group (n = 5). B The share of GFP+ leukemia cell in bone marrow have been detected by stream cytometric evaluation (n = 3). C Immature cells from the bone marrow have been checked utilizing Wright’s stain (Left), and spleen and liver infiltration have been analyzed by H&E staining (Proper). The consultant photos have been proven. D The relative expression of CD36 and PPAR-γ in GFP+ leukemia cells have been decided by qRT-PCR (n = 3). E Consultant photos of Oil Purple O staining. F The schema chart of MLL-AF9-induced leukemia was proven (n = 3). G The share of GFP+ leukemia cell in peripheral blood have been detected by stream cytometric evaluation (n = 3). H Kaplan-Meier evaluation of the survival curves of the mice in every group (n = 7). I Immature cells from the bone marrow have been checked utilizing Wright’s stain (Left), and spleen and liver infiltration have been analyzed by H&E staining (Proper). The consultant photos have been proven. J The proportion of LSCs in bone marrow was detected by stream cytometric evaluation (n = 3). Okay The relative expression of CD36 and PPAR-γ in GFP+ leukemia cells have been decided by qRT-PCR (n = 3). L Consultant photos of Oil Purple O staining. M The proportion of CD8+ T cells and NK cells in bone marrow have been decided by stream cytometric evaluation (n = 3). *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001; ns, not vital.
As LFMRS was carefully associated to the immunosuppressive state of AML (Fig. 2O), we additional discover whether or not LGALS1 regulates the immune impact of immune cells (macrophage, CD8+ T cells, and NK cells) in vivo by utilizing the MLL-AF9 retroviral transduction system (Fig. 6F). LSK cells transduced with MLL-AF9 virus from wildtype C57/B6L mice, then injected into unirradiated mice through tail vein. As proven in Fig. 6G–L, remedy with OTX008 inhibited leukemogenesis (Fig. 6G), promoted survival time (Fig. 6H), diminished leukemic cells within the bone marrow (Fig. 6I), curbed liver and splenic infiltration (Fig. 6I), and decreased LSC frequency within the bone marrow through stream evaluation (Fig. 6J), and diminished CD36, PPAR-γ, and lipid droplet ranges (Fig. 6K, L). In the meantime, we detected the immune cell counts, and located that remedy with OTX008 result in a lower within the counts of CD8+ T cells, and NK cells (Fig. 6M). Generally, LGALS1 enhances lipid metabolism reprogramming, an immunosuppressive microenvironment, and AML development in vivo.